
Sanguibacter keddieii (strain ATCC 51767 / DSM 10542 / NCFB 3025 / ST-74)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Sanguibacteraceae; Sanguibacter; Sanguibacter keddieii
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
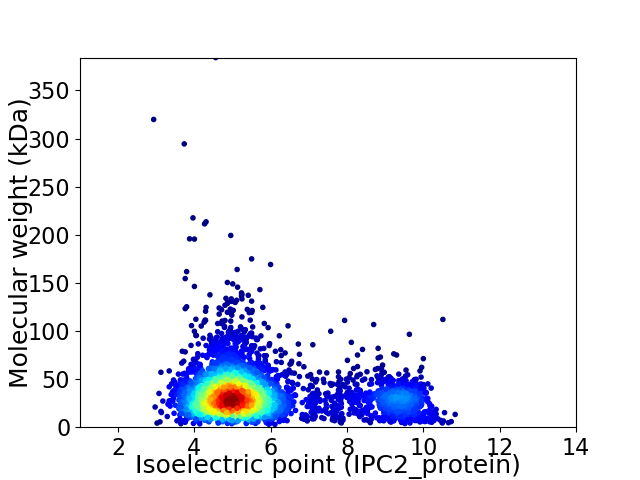
Virtual 2D-PAGE plot for 3710 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D1BDW0|D1BDW0_SANKS Assimilatory sulfite reductase (ferredoxin) OS=Sanguibacter keddieii (strain ATCC 51767 / DSM 10542 / NCFB 3025 / ST-74) OX=446469 GN=Sked_32860 PE=4 SV=1
MM1 pKa = 8.26PDD3 pKa = 3.33PNLPAGAAQPPRR15 pKa = 11.84PSARR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84LVRR25 pKa = 11.84AAAAVAGLSLVLAACSSDD43 pKa = 3.15PGTKK47 pKa = 9.86VQTGVDD53 pKa = 3.91GSSPSAGADD62 pKa = 3.41PSDD65 pKa = 4.26GASSAAPSTDD75 pKa = 3.51GIEE78 pKa = 4.64ADD80 pKa = 4.0LLPFYY85 pKa = 9.87EE86 pKa = 4.75QTVEE90 pKa = 3.83WSDD93 pKa = 3.52CGSGFEE99 pKa = 4.47CADD102 pKa = 3.22IEE104 pKa = 5.71VPFDD108 pKa = 3.62WDD110 pKa = 4.03DD111 pKa = 3.7PAADD115 pKa = 4.65TITLGAKK122 pKa = 9.75RR123 pKa = 11.84YY124 pKa = 9.29LSSGEE129 pKa = 3.92RR130 pKa = 11.84KK131 pKa = 8.49GTVLINPGGPGGSGVEE147 pKa = 4.22FVQYY151 pKa = 11.04VPFIFGKK158 pKa = 9.76PLLEE162 pKa = 4.34EE163 pKa = 4.21FDD165 pKa = 3.89VLGFDD170 pKa = 3.66PRR172 pKa = 11.84GVGEE176 pKa = 4.36SSPVACLSDD185 pKa = 3.58AEE187 pKa = 4.63RR188 pKa = 11.84DD189 pKa = 3.57VSDD192 pKa = 3.86AKK194 pKa = 10.48TYY196 pKa = 10.31TPDD199 pKa = 3.65DD200 pKa = 4.06AGLAEE205 pKa = 4.25MQADD209 pKa = 3.63AKK211 pKa = 11.3AFGEE215 pKa = 4.25KK216 pKa = 10.05CLEE219 pKa = 3.96NTGPVLGEE227 pKa = 3.92VDD229 pKa = 3.9TQSSARR235 pKa = 11.84DD236 pKa = 3.38MDD238 pKa = 4.39VIRR241 pKa = 11.84HH242 pKa = 4.62VVGDD246 pKa = 3.65AQLNYY251 pKa = 10.73LGFSYY256 pKa = 8.7GTQLGATYY264 pKa = 10.75AGLYY268 pKa = 8.48PEE270 pKa = 4.36NVGRR274 pKa = 11.84MVLDD278 pKa = 3.67GAIDD282 pKa = 3.79LRR284 pKa = 11.84LTDD287 pKa = 4.88HH288 pKa = 6.83EE289 pKa = 4.81MSLQQAVGFEE299 pKa = 4.09NALTAFVEE307 pKa = 4.49DD308 pKa = 4.42CLAGSGCPLTGTTDD322 pKa = 3.13EE323 pKa = 5.21ALAQVTGLLDD333 pKa = 3.65QILANPMPTGDD344 pKa = 3.75PARR347 pKa = 11.84PLTQTLAFYY356 pKa = 10.78GIAQPLYY363 pKa = 11.06SEE365 pKa = 5.3LLWPQLTAALDD376 pKa = 3.74AAISDD381 pKa = 4.05GDD383 pKa = 4.01GSGLLASADD392 pKa = 3.9SYY394 pKa = 11.85NSRR397 pKa = 11.84NSDD400 pKa = 2.49GTYY403 pKa = 10.78ADD405 pKa = 3.65NQGEE409 pKa = 4.26AFRR412 pKa = 11.84AINCLDD418 pKa = 3.58SRR420 pKa = 11.84SDD422 pKa = 3.54ADD424 pKa = 3.71MATLDD429 pKa = 3.96AEE431 pKa = 4.38ADD433 pKa = 4.28EE434 pKa = 4.99IVAAAPTVGEE444 pKa = 4.04FFTYY448 pKa = 10.5GGLGCADD455 pKa = 3.4WPFPQAEE462 pKa = 4.39KK463 pKa = 10.25TGDD466 pKa = 3.58LAASGAAPILVIGTTNDD483 pKa = 3.39PATPYY488 pKa = 10.5VWAQGLAEE496 pKa = 3.94QLEE499 pKa = 4.66SGVLVTYY506 pKa = 10.27EE507 pKa = 4.5GEE509 pKa = 3.9GHH511 pKa = 5.47TAYY514 pKa = 10.2GQSNSCVLDD523 pKa = 3.51TVDD526 pKa = 5.94AYY528 pKa = 10.16FTDD531 pKa = 3.8GTVPDD536 pKa = 4.43EE537 pKa = 4.61GLVCC541 pKa = 5.22
MM1 pKa = 8.26PDD3 pKa = 3.33PNLPAGAAQPPRR15 pKa = 11.84PSARR19 pKa = 11.84RR20 pKa = 11.84SRR22 pKa = 11.84LVRR25 pKa = 11.84AAAAVAGLSLVLAACSSDD43 pKa = 3.15PGTKK47 pKa = 9.86VQTGVDD53 pKa = 3.91GSSPSAGADD62 pKa = 3.41PSDD65 pKa = 4.26GASSAAPSTDD75 pKa = 3.51GIEE78 pKa = 4.64ADD80 pKa = 4.0LLPFYY85 pKa = 9.87EE86 pKa = 4.75QTVEE90 pKa = 3.83WSDD93 pKa = 3.52CGSGFEE99 pKa = 4.47CADD102 pKa = 3.22IEE104 pKa = 5.71VPFDD108 pKa = 3.62WDD110 pKa = 4.03DD111 pKa = 3.7PAADD115 pKa = 4.65TITLGAKK122 pKa = 9.75RR123 pKa = 11.84YY124 pKa = 9.29LSSGEE129 pKa = 3.92RR130 pKa = 11.84KK131 pKa = 8.49GTVLINPGGPGGSGVEE147 pKa = 4.22FVQYY151 pKa = 11.04VPFIFGKK158 pKa = 9.76PLLEE162 pKa = 4.34EE163 pKa = 4.21FDD165 pKa = 3.89VLGFDD170 pKa = 3.66PRR172 pKa = 11.84GVGEE176 pKa = 4.36SSPVACLSDD185 pKa = 3.58AEE187 pKa = 4.63RR188 pKa = 11.84DD189 pKa = 3.57VSDD192 pKa = 3.86AKK194 pKa = 10.48TYY196 pKa = 10.31TPDD199 pKa = 3.65DD200 pKa = 4.06AGLAEE205 pKa = 4.25MQADD209 pKa = 3.63AKK211 pKa = 11.3AFGEE215 pKa = 4.25KK216 pKa = 10.05CLEE219 pKa = 3.96NTGPVLGEE227 pKa = 3.92VDD229 pKa = 3.9TQSSARR235 pKa = 11.84DD236 pKa = 3.38MDD238 pKa = 4.39VIRR241 pKa = 11.84HH242 pKa = 4.62VVGDD246 pKa = 3.65AQLNYY251 pKa = 10.73LGFSYY256 pKa = 8.7GTQLGATYY264 pKa = 10.75AGLYY268 pKa = 8.48PEE270 pKa = 4.36NVGRR274 pKa = 11.84MVLDD278 pKa = 3.67GAIDD282 pKa = 3.79LRR284 pKa = 11.84LTDD287 pKa = 4.88HH288 pKa = 6.83EE289 pKa = 4.81MSLQQAVGFEE299 pKa = 4.09NALTAFVEE307 pKa = 4.49DD308 pKa = 4.42CLAGSGCPLTGTTDD322 pKa = 3.13EE323 pKa = 5.21ALAQVTGLLDD333 pKa = 3.65QILANPMPTGDD344 pKa = 3.75PARR347 pKa = 11.84PLTQTLAFYY356 pKa = 10.78GIAQPLYY363 pKa = 11.06SEE365 pKa = 5.3LLWPQLTAALDD376 pKa = 3.74AAISDD381 pKa = 4.05GDD383 pKa = 4.01GSGLLASADD392 pKa = 3.9SYY394 pKa = 11.85NSRR397 pKa = 11.84NSDD400 pKa = 2.49GTYY403 pKa = 10.78ADD405 pKa = 3.65NQGEE409 pKa = 4.26AFRR412 pKa = 11.84AINCLDD418 pKa = 3.58SRR420 pKa = 11.84SDD422 pKa = 3.54ADD424 pKa = 3.71MATLDD429 pKa = 3.96AEE431 pKa = 4.38ADD433 pKa = 4.28EE434 pKa = 4.99IVAAAPTVGEE444 pKa = 4.04FFTYY448 pKa = 10.5GGLGCADD455 pKa = 3.4WPFPQAEE462 pKa = 4.39KK463 pKa = 10.25TGDD466 pKa = 3.58LAASGAAPILVIGTTNDD483 pKa = 3.39PATPYY488 pKa = 10.5VWAQGLAEE496 pKa = 3.94QLEE499 pKa = 4.66SGVLVTYY506 pKa = 10.27EE507 pKa = 4.5GEE509 pKa = 3.9GHH511 pKa = 5.47TAYY514 pKa = 10.2GQSNSCVLDD523 pKa = 3.51TVDD526 pKa = 5.94AYY528 pKa = 10.16FTDD531 pKa = 3.8GTVPDD536 pKa = 4.43EE537 pKa = 4.61GLVCC541 pKa = 5.22
Molecular weight: 56.09 kDa
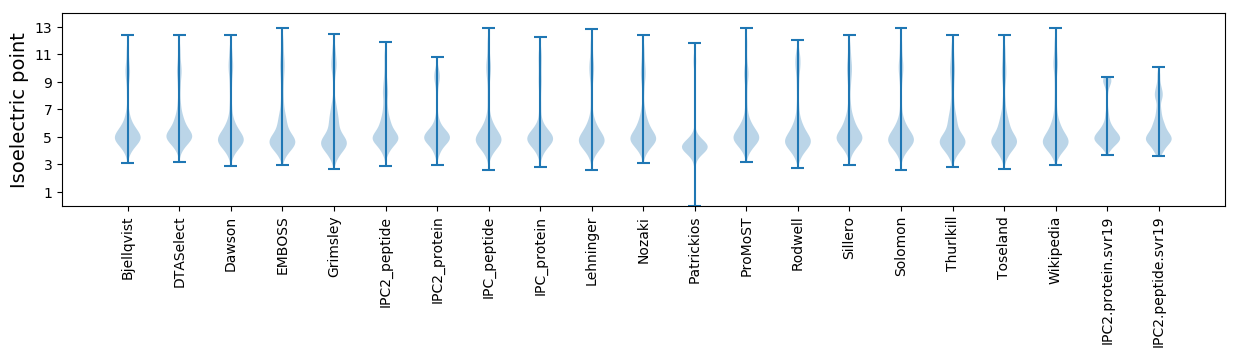
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D1BH02|D1BH02_SANKS Bifunctional protein PyrR OS=Sanguibacter keddieii (strain ATCC 51767 / DSM 10542 / NCFB 3025 / ST-74) OX=446469 GN=pyrR PE=3 SV=1
MM1 pKa = 7.02SAVRR5 pKa = 11.84RR6 pKa = 11.84LVARR10 pKa = 11.84IDD12 pKa = 3.68LFIVSLLLAVAVASFFPVRR31 pKa = 11.84GQAAEE36 pKa = 4.09GLDD39 pKa = 3.65VAVVIAVGWLFFLYY53 pKa = 10.13GVRR56 pKa = 11.84LPTKK60 pKa = 9.31EE61 pKa = 3.93AVNALRR67 pKa = 11.84DD68 pKa = 3.61WRR70 pKa = 11.84LHH72 pKa = 5.42GSVLAVTFVVFPLLGLAMQLVPSSVVPTDD101 pKa = 3.84LKK103 pKa = 11.18LGLLFLTLLPSTVQSSIAFTSIARR127 pKa = 11.84GNVAGAVVAASFSNLAGVLLTPLLVAGLLGGTVGFSADD165 pKa = 3.71SIGKK169 pKa = 9.61IGLQLLAPFVAGQLLRR185 pKa = 11.84RR186 pKa = 11.84WLAPFVLRR194 pKa = 11.84HH195 pKa = 5.61KK196 pKa = 10.61RR197 pKa = 11.84VTTLTDD203 pKa = 2.96RR204 pKa = 11.84GSVVLVVYY212 pKa = 10.24AAFSHH217 pKa = 6.14GVVDD221 pKa = 5.95GIWTTVDD228 pKa = 2.83VGSIVAVAVLCCLLLAAVLGLTTFAGKK255 pKa = 10.49RR256 pKa = 11.84FGFSRR261 pKa = 11.84ADD263 pKa = 3.46RR264 pKa = 11.84VVLLMCGSKK273 pKa = 10.36KK274 pKa = 10.56SLASGVPMATVLLPAATIGFMVLPLMIFHH303 pKa = 6.86QIQLIVCAVIARR315 pKa = 11.84RR316 pKa = 11.84MGDD319 pKa = 3.13AAEE322 pKa = 4.65PEE324 pKa = 4.32PARR327 pKa = 11.84PP328 pKa = 3.52
MM1 pKa = 7.02SAVRR5 pKa = 11.84RR6 pKa = 11.84LVARR10 pKa = 11.84IDD12 pKa = 3.68LFIVSLLLAVAVASFFPVRR31 pKa = 11.84GQAAEE36 pKa = 4.09GLDD39 pKa = 3.65VAVVIAVGWLFFLYY53 pKa = 10.13GVRR56 pKa = 11.84LPTKK60 pKa = 9.31EE61 pKa = 3.93AVNALRR67 pKa = 11.84DD68 pKa = 3.61WRR70 pKa = 11.84LHH72 pKa = 5.42GSVLAVTFVVFPLLGLAMQLVPSSVVPTDD101 pKa = 3.84LKK103 pKa = 11.18LGLLFLTLLPSTVQSSIAFTSIARR127 pKa = 11.84GNVAGAVVAASFSNLAGVLLTPLLVAGLLGGTVGFSADD165 pKa = 3.71SIGKK169 pKa = 9.61IGLQLLAPFVAGQLLRR185 pKa = 11.84RR186 pKa = 11.84WLAPFVLRR194 pKa = 11.84HH195 pKa = 5.61KK196 pKa = 10.61RR197 pKa = 11.84VTTLTDD203 pKa = 2.96RR204 pKa = 11.84GSVVLVVYY212 pKa = 10.24AAFSHH217 pKa = 6.14GVVDD221 pKa = 5.95GIWTTVDD228 pKa = 2.83VGSIVAVAVLCCLLLAAVLGLTTFAGKK255 pKa = 10.49RR256 pKa = 11.84FGFSRR261 pKa = 11.84ADD263 pKa = 3.46RR264 pKa = 11.84VVLLMCGSKK273 pKa = 10.36KK274 pKa = 10.56SLASGVPMATVLLPAATIGFMVLPLMIFHH303 pKa = 6.86QIQLIVCAVIARR315 pKa = 11.84RR316 pKa = 11.84MGDD319 pKa = 3.13AAEE322 pKa = 4.65PEE324 pKa = 4.32PARR327 pKa = 11.84PP328 pKa = 3.52
Molecular weight: 34.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1274206 |
27 |
3648 |
343.5 |
36.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.484 ± 0.049 | 0.516 ± 0.009 |
6.518 ± 0.037 | 5.551 ± 0.038 |
2.613 ± 0.026 | 9.13 ± 0.042 |
2.019 ± 0.018 | 3.148 ± 0.031 |
1.531 ± 0.026 | 10.206 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.643 ± 0.016 | 1.511 ± 0.021 |
5.734 ± 0.039 | 2.836 ± 0.021 |
7.122 ± 0.041 | 5.979 ± 0.03 |
6.987 ± 0.054 | 10.202 ± 0.049 |
1.448 ± 0.017 | 1.822 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
