
Aurantimonas sp. 22II-16-19i
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Aurantimonas; unclassified Aurantimonas
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
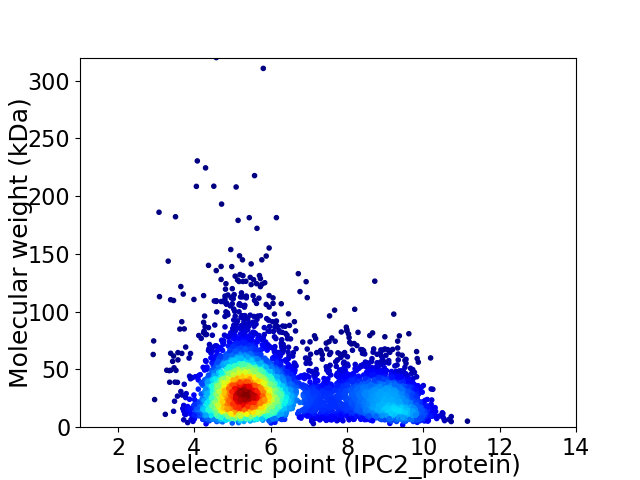
Virtual 2D-PAGE plot for 5291 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X0SV04|A0A1X0SV04_9RHIZ Translation initiation factor IF-2 (Fragment) OS=Aurantimonas sp. 22II-16-19i OX=1317114 GN=infB PE=4 SV=1
MM1 pKa = 7.3VFNIHH6 pKa = 6.31YY7 pKa = 10.41DD8 pKa = 3.34VAAFNNLAGLLSIQVLEE25 pKa = 4.58PEE27 pKa = 4.37NNADD31 pKa = 3.56SGLNGLRR38 pKa = 11.84GSAAFNFFNEE48 pKa = 4.16LGVPIEE54 pKa = 4.93DD55 pKa = 4.3GPLGALGIFVEE66 pKa = 5.06GPGDD70 pKa = 3.59IDD72 pKa = 5.45DD73 pKa = 4.95PATDD77 pKa = 3.96VYY79 pKa = 9.65HH80 pKa = 5.14TTYY83 pKa = 11.25AHH85 pKa = 4.52VHH87 pKa = 4.88GVEE90 pKa = 4.31EE91 pKa = 4.42NLTDD95 pKa = 3.75TPLGTNTFDD104 pKa = 3.23SGLAITPGEE113 pKa = 4.01YY114 pKa = 10.23LAPNVITVNGRR125 pKa = 11.84IPTFATGEE133 pKa = 3.86LWQPVTLHH141 pKa = 5.16QRR143 pKa = 11.84QVIDD147 pKa = 3.34KK148 pKa = 10.4DD149 pKa = 3.91DD150 pKa = 3.66YY151 pKa = 10.22FTINFYY157 pKa = 10.92VKK159 pKa = 9.94PGAVQQVPGQKK170 pKa = 9.49PVASANFSTSYY181 pKa = 11.07SLSTDD186 pKa = 3.46ALPHH190 pKa = 5.15YY191 pKa = 10.58AFYY194 pKa = 11.54ANMFDD199 pKa = 4.22PGVGLNSLGDD209 pKa = 3.29ILFYY213 pKa = 10.9SEE215 pKa = 4.54NSIQIGYY222 pKa = 10.47DD223 pKa = 3.39DD224 pKa = 5.2GDD226 pKa = 3.45QGNYY230 pKa = 9.08TIYY233 pKa = 10.43TGDD236 pKa = 3.42FKK238 pKa = 11.43FEE240 pKa = 4.13EE241 pKa = 4.78DD242 pKa = 3.46ASTGNVFLVEE252 pKa = 4.11GFISQIDD259 pKa = 3.6HH260 pKa = 5.84QYY262 pKa = 10.05YY263 pKa = 8.03YY264 pKa = 10.43HH265 pKa = 7.84DD266 pKa = 3.93YY267 pKa = 10.43SVTPEE272 pKa = 3.93VYY274 pKa = 10.73YY275 pKa = 11.01SGLWTASSVGITAAEE290 pKa = 3.85ARR292 pKa = 11.84DD293 pKa = 4.11LFGQDD298 pKa = 2.83FSEE301 pKa = 5.27RR302 pKa = 11.84IFRR305 pKa = 11.84GNDD308 pKa = 3.06TLKK311 pKa = 11.02GSTEE315 pKa = 3.95ADD317 pKa = 3.24RR318 pKa = 11.84LFAFAGNDD326 pKa = 3.27ILIGGAGADD335 pKa = 3.63QLNGGTGSDD344 pKa = 3.31TASYY348 pKa = 10.59AGSLSVRR355 pKa = 11.84VNLTNGRR362 pKa = 11.84GSFGDD367 pKa = 3.5AEE369 pKa = 4.65GDD371 pKa = 3.67VLIGIEE377 pKa = 4.42NITGSEE383 pKa = 4.05RR384 pKa = 11.84ADD386 pKa = 3.41TLLGNSGANVINGEE400 pKa = 4.07GGFDD404 pKa = 4.1EE405 pKa = 4.12IHH407 pKa = 6.83GYY409 pKa = 10.2AGNDD413 pKa = 2.96RR414 pKa = 11.84LYY416 pKa = 11.2GGNGLDD422 pKa = 4.45KK423 pKa = 11.08LYY425 pKa = 11.39GDD427 pKa = 5.4FNADD431 pKa = 3.41MLFGGAGGDD440 pKa = 3.88TLSGGAGPDD449 pKa = 3.14KK450 pKa = 11.25VYY452 pKa = 11.27GEE454 pKa = 4.64SGNDD458 pKa = 3.63NIVGDD463 pKa = 4.26ADD465 pKa = 3.72GSEE468 pKa = 4.02DD469 pKa = 3.96VYY471 pKa = 11.77DD472 pKa = 4.59GGTEE476 pKa = 3.88IDD478 pKa = 3.72TVDD481 pKa = 3.52YY482 pKa = 10.7GAVASAINVNLSLNFAMGAPIGTDD506 pKa = 2.79RR507 pKa = 11.84IFAVEE512 pKa = 4.87RR513 pKa = 11.84ILAGAGNDD521 pKa = 3.61KK522 pKa = 9.95LTGNAFTTLLSGAGGNDD539 pKa = 3.5VIVGGIGDD547 pKa = 3.74NKK549 pKa = 10.68LYY551 pKa = 11.11GGDD554 pKa = 3.19GHH556 pKa = 7.52DD557 pKa = 4.11NLRR560 pKa = 11.84GLAGFDD566 pKa = 3.2QFFGGNGNDD575 pKa = 3.55RR576 pKa = 11.84MQGDD580 pKa = 4.49FNADD584 pKa = 3.12TFIFANGFGRR594 pKa = 11.84DD595 pKa = 3.5VIADD599 pKa = 4.12FEE601 pKa = 4.48ALNAAEE607 pKa = 4.93QIDD610 pKa = 4.35LSGVTNITSFADD622 pKa = 3.75LAANHH627 pKa = 6.87LSQVGGNAVITDD639 pKa = 4.21GFNTITLNGVAIGDD653 pKa = 4.18LDD655 pKa = 3.57ATDD658 pKa = 5.74FIFF661 pKa = 5.37
MM1 pKa = 7.3VFNIHH6 pKa = 6.31YY7 pKa = 10.41DD8 pKa = 3.34VAAFNNLAGLLSIQVLEE25 pKa = 4.58PEE27 pKa = 4.37NNADD31 pKa = 3.56SGLNGLRR38 pKa = 11.84GSAAFNFFNEE48 pKa = 4.16LGVPIEE54 pKa = 4.93DD55 pKa = 4.3GPLGALGIFVEE66 pKa = 5.06GPGDD70 pKa = 3.59IDD72 pKa = 5.45DD73 pKa = 4.95PATDD77 pKa = 3.96VYY79 pKa = 9.65HH80 pKa = 5.14TTYY83 pKa = 11.25AHH85 pKa = 4.52VHH87 pKa = 4.88GVEE90 pKa = 4.31EE91 pKa = 4.42NLTDD95 pKa = 3.75TPLGTNTFDD104 pKa = 3.23SGLAITPGEE113 pKa = 4.01YY114 pKa = 10.23LAPNVITVNGRR125 pKa = 11.84IPTFATGEE133 pKa = 3.86LWQPVTLHH141 pKa = 5.16QRR143 pKa = 11.84QVIDD147 pKa = 3.34KK148 pKa = 10.4DD149 pKa = 3.91DD150 pKa = 3.66YY151 pKa = 10.22FTINFYY157 pKa = 10.92VKK159 pKa = 9.94PGAVQQVPGQKK170 pKa = 9.49PVASANFSTSYY181 pKa = 11.07SLSTDD186 pKa = 3.46ALPHH190 pKa = 5.15YY191 pKa = 10.58AFYY194 pKa = 11.54ANMFDD199 pKa = 4.22PGVGLNSLGDD209 pKa = 3.29ILFYY213 pKa = 10.9SEE215 pKa = 4.54NSIQIGYY222 pKa = 10.47DD223 pKa = 3.39DD224 pKa = 5.2GDD226 pKa = 3.45QGNYY230 pKa = 9.08TIYY233 pKa = 10.43TGDD236 pKa = 3.42FKK238 pKa = 11.43FEE240 pKa = 4.13EE241 pKa = 4.78DD242 pKa = 3.46ASTGNVFLVEE252 pKa = 4.11GFISQIDD259 pKa = 3.6HH260 pKa = 5.84QYY262 pKa = 10.05YY263 pKa = 8.03YY264 pKa = 10.43HH265 pKa = 7.84DD266 pKa = 3.93YY267 pKa = 10.43SVTPEE272 pKa = 3.93VYY274 pKa = 10.73YY275 pKa = 11.01SGLWTASSVGITAAEE290 pKa = 3.85ARR292 pKa = 11.84DD293 pKa = 4.11LFGQDD298 pKa = 2.83FSEE301 pKa = 5.27RR302 pKa = 11.84IFRR305 pKa = 11.84GNDD308 pKa = 3.06TLKK311 pKa = 11.02GSTEE315 pKa = 3.95ADD317 pKa = 3.24RR318 pKa = 11.84LFAFAGNDD326 pKa = 3.27ILIGGAGADD335 pKa = 3.63QLNGGTGSDD344 pKa = 3.31TASYY348 pKa = 10.59AGSLSVRR355 pKa = 11.84VNLTNGRR362 pKa = 11.84GSFGDD367 pKa = 3.5AEE369 pKa = 4.65GDD371 pKa = 3.67VLIGIEE377 pKa = 4.42NITGSEE383 pKa = 4.05RR384 pKa = 11.84ADD386 pKa = 3.41TLLGNSGANVINGEE400 pKa = 4.07GGFDD404 pKa = 4.1EE405 pKa = 4.12IHH407 pKa = 6.83GYY409 pKa = 10.2AGNDD413 pKa = 2.96RR414 pKa = 11.84LYY416 pKa = 11.2GGNGLDD422 pKa = 4.45KK423 pKa = 11.08LYY425 pKa = 11.39GDD427 pKa = 5.4FNADD431 pKa = 3.41MLFGGAGGDD440 pKa = 3.88TLSGGAGPDD449 pKa = 3.14KK450 pKa = 11.25VYY452 pKa = 11.27GEE454 pKa = 4.64SGNDD458 pKa = 3.63NIVGDD463 pKa = 4.26ADD465 pKa = 3.72GSEE468 pKa = 4.02DD469 pKa = 3.96VYY471 pKa = 11.77DD472 pKa = 4.59GGTEE476 pKa = 3.88IDD478 pKa = 3.72TVDD481 pKa = 3.52YY482 pKa = 10.7GAVASAINVNLSLNFAMGAPIGTDD506 pKa = 2.79RR507 pKa = 11.84IFAVEE512 pKa = 4.87RR513 pKa = 11.84ILAGAGNDD521 pKa = 3.61KK522 pKa = 9.95LTGNAFTTLLSGAGGNDD539 pKa = 3.5VIVGGIGDD547 pKa = 3.74NKK549 pKa = 10.68LYY551 pKa = 11.11GGDD554 pKa = 3.19GHH556 pKa = 7.52DD557 pKa = 4.11NLRR560 pKa = 11.84GLAGFDD566 pKa = 3.2QFFGGNGNDD575 pKa = 3.55RR576 pKa = 11.84MQGDD580 pKa = 4.49FNADD584 pKa = 3.12TFIFANGFGRR594 pKa = 11.84DD595 pKa = 3.5VIADD599 pKa = 4.12FEE601 pKa = 4.48ALNAAEE607 pKa = 4.93QIDD610 pKa = 4.35LSGVTNITSFADD622 pKa = 3.75LAANHH627 pKa = 6.87LSQVGGNAVITDD639 pKa = 4.21GFNTITLNGVAIGDD653 pKa = 4.18LDD655 pKa = 3.57ATDD658 pKa = 5.74FIFF661 pKa = 5.37
Molecular weight: 69.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X0T213|A0A1X0T213_9RHIZ Putative major facilitator superfamily MFS-1 OS=Aurantimonas sp. 22II-16-19i OX=1317114 GN=ATO4_13590 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33SGQKK29 pKa = 9.93VIAARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.33SGQKK29 pKa = 9.93VIAARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1651721 |
17 |
3005 |
312.2 |
33.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.287 ± 0.06 | 0.788 ± 0.01 |
5.905 ± 0.032 | 6.004 ± 0.031 |
3.752 ± 0.025 | 9.037 ± 0.048 |
1.86 ± 0.017 | 5.045 ± 0.028 |
2.89 ± 0.029 | 9.847 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.433 ± 0.014 | 2.295 ± 0.021 |
5.204 ± 0.032 | 2.791 ± 0.019 |
7.355 ± 0.038 | 5.429 ± 0.023 |
5.315 ± 0.025 | 7.475 ± 0.029 |
1.205 ± 0.014 | 2.084 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
