
Prosthecochloris sp. GSB1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Chlorobi; Chlorobia; Chlorobiales; Chlorobiaceae; Prosthecochloris; unclassified Prosthecochloris
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
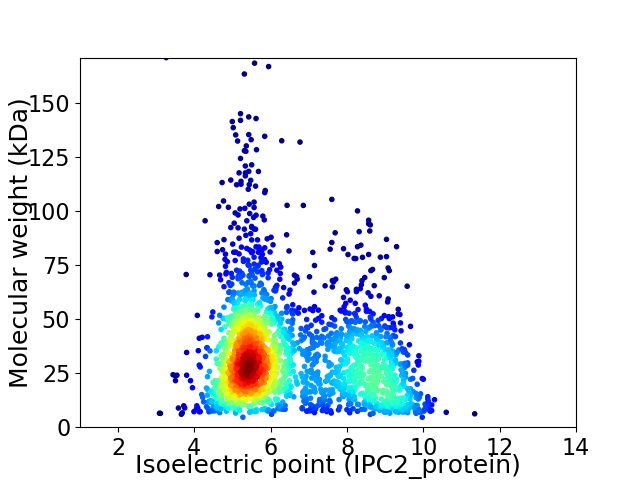
Virtual 2D-PAGE plot for 2269 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A222SIX9|A0A222SIX9_9CHLB Uncharacterized protein OS=Prosthecochloris sp. GSB1 OX=281093 GN=CHL67_08285 PE=4 SV=1
MM1 pKa = 7.91RR2 pKa = 11.84KK3 pKa = 9.86NIQHH7 pKa = 6.64KK8 pKa = 10.84LEE10 pKa = 4.84DD11 pKa = 3.48YY12 pKa = 10.5FLRR15 pKa = 11.84EE16 pKa = 3.59RR17 pKa = 11.84YY18 pKa = 9.63LPEE21 pKa = 3.93EE22 pKa = 4.0SQEE25 pKa = 3.96FLKK28 pKa = 11.06DD29 pKa = 3.26SAEE32 pKa = 4.24YY33 pKa = 10.31AVPATDD39 pKa = 3.39SASAGTAIATGYY51 pKa = 8.3KK52 pKa = 9.2TDD54 pKa = 4.0AGNISWASGDD64 pKa = 3.86PEE66 pKa = 4.42NGEE69 pKa = 4.12LTTIAEE75 pKa = 4.51TLRR78 pKa = 11.84SEE80 pKa = 3.95EE81 pKa = 4.0GFAIGVVSTVPFSHH95 pKa = 6.69ATPAVFVSHH104 pKa = 7.18DD105 pKa = 3.56VNRR108 pKa = 11.84GNKK111 pKa = 9.23YY112 pKa = 10.72DD113 pKa = 3.59IAHH116 pKa = 7.3EE117 pKa = 4.25ILFEE121 pKa = 4.11TQPDD125 pKa = 3.92VVIGSGYY132 pKa = 8.68EE133 pKa = 3.54NSYY136 pKa = 10.17FARR139 pKa = 11.84TKK141 pKa = 10.34TVDD144 pKa = 3.39EE145 pKa = 4.93NGNDD149 pKa = 3.55YY150 pKa = 11.45NDD152 pKa = 5.07DD153 pKa = 3.32YY154 pKa = 12.02DD155 pKa = 4.59SFVNGTDD162 pKa = 3.0GTDD165 pKa = 3.34YY166 pKa = 11.44VFVQRR171 pKa = 11.84DD172 pKa = 3.32EE173 pKa = 5.2DD174 pKa = 4.49LDD176 pKa = 4.93GGDD179 pKa = 5.16LLMEE183 pKa = 4.97AANSVDD189 pKa = 3.61LQAGEE194 pKa = 4.38KK195 pKa = 10.69LFGLFGTSGGNFEE208 pKa = 5.16YY209 pKa = 11.14YY210 pKa = 10.41DD211 pKa = 3.64VDD213 pKa = 4.65DD214 pKa = 5.12NPGSPSITRR223 pKa = 11.84DD224 pKa = 3.01GDD226 pKa = 3.64GDD228 pKa = 4.22GVDD231 pKa = 4.33EE232 pKa = 5.69DD233 pKa = 4.19PTLAEE238 pKa = 3.96ATIAALTVLDD248 pKa = 3.74QDD250 pKa = 3.36EE251 pKa = 5.26DD252 pKa = 4.41GFFVMIEE259 pKa = 3.91QGDD262 pKa = 3.91IDD264 pKa = 4.49WNNHH268 pKa = 4.51ANDD271 pKa = 4.04YY272 pKa = 10.79EE273 pKa = 4.29NMIGSVYY280 pKa = 10.98DD281 pKa = 3.59LDD283 pKa = 3.66QAVRR287 pKa = 11.84VVEE290 pKa = 4.37AYY292 pKa = 10.9VDD294 pKa = 3.68IEE296 pKa = 4.71GDD298 pKa = 3.89DD299 pKa = 3.97MDD301 pKa = 3.81WSNTLVIVTSDD312 pKa = 3.02HH313 pKa = 6.5SNSYY317 pKa = 9.37MRR319 pKa = 11.84TNEE322 pKa = 3.68EE323 pKa = 4.41LGIGDD328 pKa = 4.99LPSQEE333 pKa = 3.7GSPYY337 pKa = 11.15DD338 pKa = 3.32FTYY341 pKa = 10.98PNGEE345 pKa = 4.0VTYY348 pKa = 8.57GTTGHH353 pKa = 5.87TNEE356 pKa = 4.89LVTLQARR363 pKa = 11.84GAGAEE368 pKa = 4.09LFEE371 pKa = 5.14EE372 pKa = 5.08YY373 pKa = 10.54AGSWYY378 pKa = 10.13EE379 pKa = 3.89GTSIVDD385 pKa = 3.03NTQIYY390 pKa = 10.96DD391 pKa = 3.51VMLRR395 pKa = 11.84AAQEE399 pKa = 3.97EE400 pKa = 4.96GVEE403 pKa = 4.14HH404 pKa = 6.77VILFIGDD411 pKa = 3.77GMNVEE416 pKa = 4.87HH417 pKa = 6.89EE418 pKa = 4.46MAASRR423 pKa = 11.84YY424 pKa = 10.08LYY426 pKa = 11.08GDD428 pKa = 4.11DD429 pKa = 5.86QSLAWDD435 pKa = 3.35DD436 pKa = 3.54WGTEE440 pKa = 4.19EE441 pKa = 3.91IGGYY445 pKa = 9.97AGYY448 pKa = 10.76SSTWDD453 pKa = 2.8INTYY457 pKa = 9.47NRR459 pKa = 11.84YY460 pKa = 9.29ADD462 pKa = 3.64EE463 pKa = 4.55VGAEE467 pKa = 4.24AYY469 pKa = 10.45HH470 pKa = 6.3PVTFNPLVGYY480 pKa = 9.17NPKK483 pKa = 10.4RR484 pKa = 11.84GGEE487 pKa = 3.98VPYY490 pKa = 10.0PLEE493 pKa = 4.22SDD495 pKa = 3.57SFFGHH500 pKa = 6.76RR501 pKa = 11.84RR502 pKa = 11.84GQTIIGSKK510 pKa = 10.23NDD512 pKa = 3.23EE513 pKa = 4.03QLIGDD518 pKa = 4.06EE519 pKa = 4.15NDD521 pKa = 3.07NRR523 pKa = 11.84ILGLKK528 pKa = 10.01GDD530 pKa = 4.21DD531 pKa = 3.83VLNGGLGADD540 pKa = 4.11RR541 pKa = 11.84LRR543 pKa = 11.84GGNGSDD549 pKa = 3.14TFVYY553 pKa = 10.59EE554 pKa = 4.09SLKK557 pKa = 11.1DD558 pKa = 3.65SLVTPGARR566 pKa = 11.84DD567 pKa = 3.49VITDD571 pKa = 3.96FKK573 pKa = 10.86PGEE576 pKa = 4.05DD577 pKa = 4.58LIDD580 pKa = 4.08LSSLAEE586 pKa = 3.95FDD588 pKa = 4.57GVILGAGDD596 pKa = 3.53HH597 pKa = 6.81FDD599 pKa = 3.89YY600 pKa = 10.65KK601 pKa = 11.27ASGAQLLFDD610 pKa = 4.13SEE612 pKa = 4.84AGILYY617 pKa = 10.71GNVDD621 pKa = 3.65SDD623 pKa = 3.89NYY625 pKa = 11.01SDD627 pKa = 5.14FAIEE631 pKa = 4.67LIGVDD636 pKa = 3.83SLEE639 pKa = 4.63ASDD642 pKa = 5.1FMLVAA647 pKa = 5.15
MM1 pKa = 7.91RR2 pKa = 11.84KK3 pKa = 9.86NIQHH7 pKa = 6.64KK8 pKa = 10.84LEE10 pKa = 4.84DD11 pKa = 3.48YY12 pKa = 10.5FLRR15 pKa = 11.84EE16 pKa = 3.59RR17 pKa = 11.84YY18 pKa = 9.63LPEE21 pKa = 3.93EE22 pKa = 4.0SQEE25 pKa = 3.96FLKK28 pKa = 11.06DD29 pKa = 3.26SAEE32 pKa = 4.24YY33 pKa = 10.31AVPATDD39 pKa = 3.39SASAGTAIATGYY51 pKa = 8.3KK52 pKa = 9.2TDD54 pKa = 4.0AGNISWASGDD64 pKa = 3.86PEE66 pKa = 4.42NGEE69 pKa = 4.12LTTIAEE75 pKa = 4.51TLRR78 pKa = 11.84SEE80 pKa = 3.95EE81 pKa = 4.0GFAIGVVSTVPFSHH95 pKa = 6.69ATPAVFVSHH104 pKa = 7.18DD105 pKa = 3.56VNRR108 pKa = 11.84GNKK111 pKa = 9.23YY112 pKa = 10.72DD113 pKa = 3.59IAHH116 pKa = 7.3EE117 pKa = 4.25ILFEE121 pKa = 4.11TQPDD125 pKa = 3.92VVIGSGYY132 pKa = 8.68EE133 pKa = 3.54NSYY136 pKa = 10.17FARR139 pKa = 11.84TKK141 pKa = 10.34TVDD144 pKa = 3.39EE145 pKa = 4.93NGNDD149 pKa = 3.55YY150 pKa = 11.45NDD152 pKa = 5.07DD153 pKa = 3.32YY154 pKa = 12.02DD155 pKa = 4.59SFVNGTDD162 pKa = 3.0GTDD165 pKa = 3.34YY166 pKa = 11.44VFVQRR171 pKa = 11.84DD172 pKa = 3.32EE173 pKa = 5.2DD174 pKa = 4.49LDD176 pKa = 4.93GGDD179 pKa = 5.16LLMEE183 pKa = 4.97AANSVDD189 pKa = 3.61LQAGEE194 pKa = 4.38KK195 pKa = 10.69LFGLFGTSGGNFEE208 pKa = 5.16YY209 pKa = 11.14YY210 pKa = 10.41DD211 pKa = 3.64VDD213 pKa = 4.65DD214 pKa = 5.12NPGSPSITRR223 pKa = 11.84DD224 pKa = 3.01GDD226 pKa = 3.64GDD228 pKa = 4.22GVDD231 pKa = 4.33EE232 pKa = 5.69DD233 pKa = 4.19PTLAEE238 pKa = 3.96ATIAALTVLDD248 pKa = 3.74QDD250 pKa = 3.36EE251 pKa = 5.26DD252 pKa = 4.41GFFVMIEE259 pKa = 3.91QGDD262 pKa = 3.91IDD264 pKa = 4.49WNNHH268 pKa = 4.51ANDD271 pKa = 4.04YY272 pKa = 10.79EE273 pKa = 4.29NMIGSVYY280 pKa = 10.98DD281 pKa = 3.59LDD283 pKa = 3.66QAVRR287 pKa = 11.84VVEE290 pKa = 4.37AYY292 pKa = 10.9VDD294 pKa = 3.68IEE296 pKa = 4.71GDD298 pKa = 3.89DD299 pKa = 3.97MDD301 pKa = 3.81WSNTLVIVTSDD312 pKa = 3.02HH313 pKa = 6.5SNSYY317 pKa = 9.37MRR319 pKa = 11.84TNEE322 pKa = 3.68EE323 pKa = 4.41LGIGDD328 pKa = 4.99LPSQEE333 pKa = 3.7GSPYY337 pKa = 11.15DD338 pKa = 3.32FTYY341 pKa = 10.98PNGEE345 pKa = 4.0VTYY348 pKa = 8.57GTTGHH353 pKa = 5.87TNEE356 pKa = 4.89LVTLQARR363 pKa = 11.84GAGAEE368 pKa = 4.09LFEE371 pKa = 5.14EE372 pKa = 5.08YY373 pKa = 10.54AGSWYY378 pKa = 10.13EE379 pKa = 3.89GTSIVDD385 pKa = 3.03NTQIYY390 pKa = 10.96DD391 pKa = 3.51VMLRR395 pKa = 11.84AAQEE399 pKa = 3.97EE400 pKa = 4.96GVEE403 pKa = 4.14HH404 pKa = 6.77VILFIGDD411 pKa = 3.77GMNVEE416 pKa = 4.87HH417 pKa = 6.89EE418 pKa = 4.46MAASRR423 pKa = 11.84YY424 pKa = 10.08LYY426 pKa = 11.08GDD428 pKa = 4.11DD429 pKa = 5.86QSLAWDD435 pKa = 3.35DD436 pKa = 3.54WGTEE440 pKa = 4.19EE441 pKa = 3.91IGGYY445 pKa = 9.97AGYY448 pKa = 10.76SSTWDD453 pKa = 2.8INTYY457 pKa = 9.47NRR459 pKa = 11.84YY460 pKa = 9.29ADD462 pKa = 3.64EE463 pKa = 4.55VGAEE467 pKa = 4.24AYY469 pKa = 10.45HH470 pKa = 6.3PVTFNPLVGYY480 pKa = 9.17NPKK483 pKa = 10.4RR484 pKa = 11.84GGEE487 pKa = 3.98VPYY490 pKa = 10.0PLEE493 pKa = 4.22SDD495 pKa = 3.57SFFGHH500 pKa = 6.76RR501 pKa = 11.84RR502 pKa = 11.84GQTIIGSKK510 pKa = 10.23NDD512 pKa = 3.23EE513 pKa = 4.03QLIGDD518 pKa = 4.06EE519 pKa = 4.15NDD521 pKa = 3.07NRR523 pKa = 11.84ILGLKK528 pKa = 10.01GDD530 pKa = 4.21DD531 pKa = 3.83VLNGGLGADD540 pKa = 4.11RR541 pKa = 11.84LRR543 pKa = 11.84GGNGSDD549 pKa = 3.14TFVYY553 pKa = 10.59EE554 pKa = 4.09SLKK557 pKa = 11.1DD558 pKa = 3.65SLVTPGARR566 pKa = 11.84DD567 pKa = 3.49VITDD571 pKa = 3.96FKK573 pKa = 10.86PGEE576 pKa = 4.05DD577 pKa = 4.58LIDD580 pKa = 4.08LSSLAEE586 pKa = 3.95FDD588 pKa = 4.57GVILGAGDD596 pKa = 3.53HH597 pKa = 6.81FDD599 pKa = 3.89YY600 pKa = 10.65KK601 pKa = 11.27ASGAQLLFDD610 pKa = 4.13SEE612 pKa = 4.84AGILYY617 pKa = 10.71GNVDD621 pKa = 3.65SDD623 pKa = 3.89NYY625 pKa = 11.01SDD627 pKa = 5.14FAIEE631 pKa = 4.67LIGVDD636 pKa = 3.83SLEE639 pKa = 4.63ASDD642 pKa = 5.1FMLVAA647 pKa = 5.15
Molecular weight: 70.73 kDa
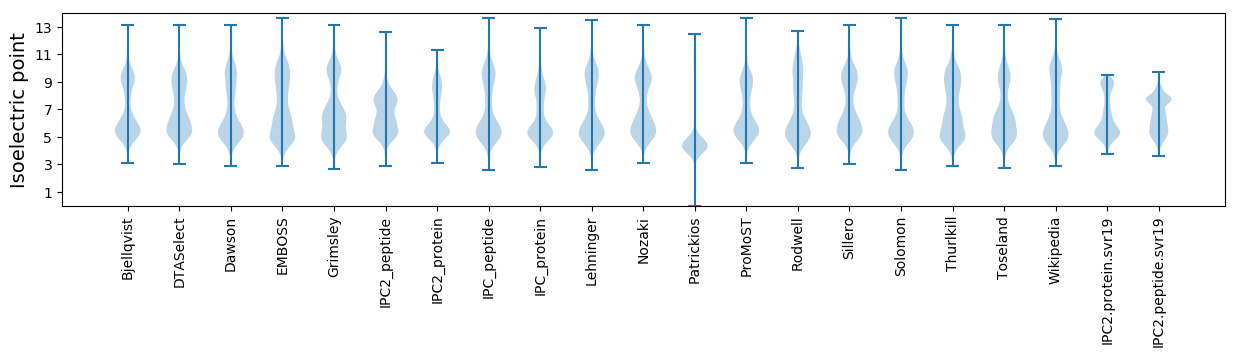
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A222SLJ2|A0A222SLJ2_9CHLB FAD:protein FMN transferase OS=Prosthecochloris sp. GSB1 OX=281093 GN=CHL67_04785 PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 3.66NRR10 pKa = 11.84KK11 pKa = 9.13RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 8.63HH16 pKa = 3.75GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84RR29 pKa = 11.84VINSRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84RR40 pKa = 11.84SLTVSSAMGTRR51 pKa = 11.84GTT53 pKa = 3.98
MM1 pKa = 7.38KK2 pKa = 9.6RR3 pKa = 11.84TFQPHH8 pKa = 3.66NRR10 pKa = 11.84KK11 pKa = 9.13RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 8.63HH16 pKa = 3.75GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84RR29 pKa = 11.84VINSRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84RR40 pKa = 11.84SLTVSSAMGTRR51 pKa = 11.84GTT53 pKa = 3.98
Molecular weight: 6.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
719611 |
38 |
1639 |
317.1 |
35.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.88 ± 0.047 | 1.146 ± 0.022 |
5.484 ± 0.036 | 7.027 ± 0.05 |
4.46 ± 0.034 | 8.039 ± 0.05 |
2.046 ± 0.02 | 5.954 ± 0.042 |
4.8 ± 0.044 | 10.059 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.564 ± 0.024 | 3.302 ± 0.024 |
4.341 ± 0.036 | 2.688 ± 0.024 |
6.757 ± 0.049 | 6.344 ± 0.035 |
4.885 ± 0.035 | 7.262 ± 0.042 |
1.048 ± 0.02 | 2.913 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
