
Cherry rusty mottle associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Quinvirinae; Robigovirus
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
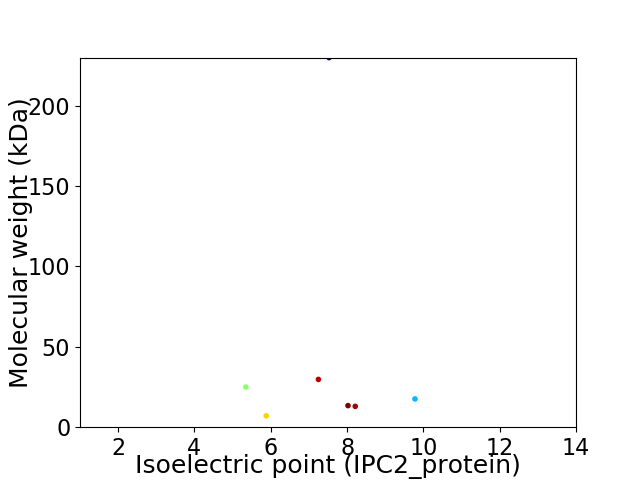
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
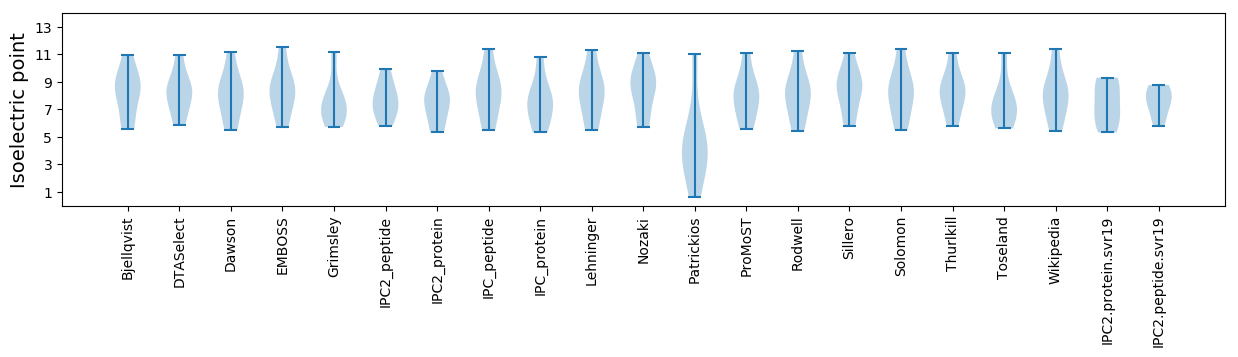
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M9QTU6|M9QTU6_9VIRU Uncharacterized protein OS=Cherry rusty mottle associated virus OX=1312929 PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 5.22IAHH5 pKa = 7.03GYY7 pKa = 9.99LLDD10 pKa = 4.16ANFEE14 pKa = 4.14RR15 pKa = 11.84TEE17 pKa = 4.1FSLSFPIVVHH27 pKa = 6.15GVPGCGKK34 pKa = 8.15STFVKK39 pKa = 10.44RR40 pKa = 11.84LLDD43 pKa = 3.91CEE45 pKa = 5.06DD46 pKa = 4.21FHH48 pKa = 7.85AQSYY52 pKa = 9.73GVVKK56 pKa = 8.74PTNLAGRR63 pKa = 11.84GVEE66 pKa = 4.54KK67 pKa = 10.53ALQPLQSGFNVLDD80 pKa = 4.47EE81 pKa = 4.39YY82 pKa = 11.6LSGPSYY88 pKa = 11.04EE89 pKa = 5.09GFDD92 pKa = 4.41LLLSDD97 pKa = 5.16PYY99 pKa = 11.53QNFRR103 pKa = 11.84KK104 pKa = 9.6PLTAHH109 pKa = 7.29FINSSTYY116 pKa = 9.19RR117 pKa = 11.84FGHH120 pKa = 5.45SVCKK124 pKa = 10.33YY125 pKa = 9.91LNLLGFEE132 pKa = 4.36INSKK136 pKa = 10.17RR137 pKa = 11.84EE138 pKa = 3.61KK139 pKa = 9.96DD140 pKa = 3.24TEE142 pKa = 4.82LIFGRR147 pKa = 11.84IFEE150 pKa = 4.28GTIKK154 pKa = 11.09GEE156 pKa = 3.99IICFEE161 pKa = 4.4KK162 pKa = 10.64EE163 pKa = 4.11VQEE166 pKa = 4.96LLDD169 pKa = 3.58NHH171 pKa = 5.8SAKK174 pKa = 10.29YY175 pKa = 9.18HH176 pKa = 5.94HH177 pKa = 7.28PCNLRR182 pKa = 11.84GAEE185 pKa = 4.08FEE187 pKa = 4.26HH188 pKa = 6.12VTFISAHH195 pKa = 6.36SDD197 pKa = 3.02LQEE200 pKa = 3.96IVGPDD205 pKa = 3.73LYY207 pKa = 11.37VALTRR212 pKa = 11.84ASEE215 pKa = 4.27SLTILTPP222 pKa = 3.99
MM1 pKa = 7.63EE2 pKa = 5.22IAHH5 pKa = 7.03GYY7 pKa = 9.99LLDD10 pKa = 4.16ANFEE14 pKa = 4.14RR15 pKa = 11.84TEE17 pKa = 4.1FSLSFPIVVHH27 pKa = 6.15GVPGCGKK34 pKa = 8.15STFVKK39 pKa = 10.44RR40 pKa = 11.84LLDD43 pKa = 3.91CEE45 pKa = 5.06DD46 pKa = 4.21FHH48 pKa = 7.85AQSYY52 pKa = 9.73GVVKK56 pKa = 8.74PTNLAGRR63 pKa = 11.84GVEE66 pKa = 4.54KK67 pKa = 10.53ALQPLQSGFNVLDD80 pKa = 4.47EE81 pKa = 4.39YY82 pKa = 11.6LSGPSYY88 pKa = 11.04EE89 pKa = 5.09GFDD92 pKa = 4.41LLLSDD97 pKa = 5.16PYY99 pKa = 11.53QNFRR103 pKa = 11.84KK104 pKa = 9.6PLTAHH109 pKa = 7.29FINSSTYY116 pKa = 9.19RR117 pKa = 11.84FGHH120 pKa = 5.45SVCKK124 pKa = 10.33YY125 pKa = 9.91LNLLGFEE132 pKa = 4.36INSKK136 pKa = 10.17RR137 pKa = 11.84EE138 pKa = 3.61KK139 pKa = 9.96DD140 pKa = 3.24TEE142 pKa = 4.82LIFGRR147 pKa = 11.84IFEE150 pKa = 4.28GTIKK154 pKa = 11.09GEE156 pKa = 3.99IICFEE161 pKa = 4.4KK162 pKa = 10.64EE163 pKa = 4.11VQEE166 pKa = 4.96LLDD169 pKa = 3.58NHH171 pKa = 5.8SAKK174 pKa = 10.29YY175 pKa = 9.18HH176 pKa = 5.94HH177 pKa = 7.28PCNLRR182 pKa = 11.84GAEE185 pKa = 4.08FEE187 pKa = 4.26HH188 pKa = 6.12VTFISAHH195 pKa = 6.36SDD197 pKa = 3.02LQEE200 pKa = 3.96IVGPDD205 pKa = 3.73LYY207 pKa = 11.37VALTRR212 pKa = 11.84ASEE215 pKa = 4.27SLTILTPP222 pKa = 3.99
Molecular weight: 24.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9QXN6|M9QXN6_9VIRU Movement protein TGBp3 OS=Cherry rusty mottle associated virus OX=1312929 PE=3 SV=1
MM1 pKa = 7.07MMEE4 pKa = 4.56PSSWTLQGKK13 pKa = 9.32RR14 pKa = 11.84SQRR17 pKa = 11.84KK18 pKa = 8.15RR19 pKa = 11.84RR20 pKa = 11.84PGRR23 pKa = 11.84TLSFLEE29 pKa = 4.4QEE31 pKa = 4.26ASNPKK36 pKa = 10.3DD37 pKa = 3.04RR38 pKa = 11.84TLRR41 pKa = 11.84SSGQEE46 pKa = 3.9EE47 pKa = 4.72EE48 pKa = 4.63EE49 pKa = 4.8LPLTPKK55 pKa = 10.26IPPLVLAEE63 pKa = 4.06TSSAVFKK70 pKa = 10.85IRR72 pKa = 11.84TQLRR76 pKa = 11.84LTLPLTTPSRR86 pKa = 11.84QLRR89 pKa = 11.84LIGSSILRR97 pKa = 11.84FQRR100 pKa = 11.84QKK102 pKa = 11.41YY103 pKa = 9.86LIASLILSGTVIITAPVTKK122 pKa = 10.31QSLLVEE128 pKa = 4.1QSVEE132 pKa = 4.08LNLKK136 pKa = 9.64VLLVLLGATALYY148 pKa = 10.3AVSARR153 pKa = 11.84NMLQQ157 pKa = 2.86
MM1 pKa = 7.07MMEE4 pKa = 4.56PSSWTLQGKK13 pKa = 9.32RR14 pKa = 11.84SQRR17 pKa = 11.84KK18 pKa = 8.15RR19 pKa = 11.84RR20 pKa = 11.84PGRR23 pKa = 11.84TLSFLEE29 pKa = 4.4QEE31 pKa = 4.26ASNPKK36 pKa = 10.3DD37 pKa = 3.04RR38 pKa = 11.84TLRR41 pKa = 11.84SSGQEE46 pKa = 3.9EE47 pKa = 4.72EE48 pKa = 4.63EE49 pKa = 4.8LPLTPKK55 pKa = 10.26IPPLVLAEE63 pKa = 4.06TSSAVFKK70 pKa = 10.85IRR72 pKa = 11.84TQLRR76 pKa = 11.84LTLPLTTPSRR86 pKa = 11.84QLRR89 pKa = 11.84LIGSSILRR97 pKa = 11.84FQRR100 pKa = 11.84QKK102 pKa = 11.41YY103 pKa = 9.86LIASLILSGTVIITAPVTKK122 pKa = 10.31QSLLVEE128 pKa = 4.1QSVEE132 pKa = 4.08LNLKK136 pKa = 9.64VLLVLLGATALYY148 pKa = 10.3AVSARR153 pKa = 11.84NMLQQ157 pKa = 2.86
Molecular weight: 17.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2977 |
67 |
2027 |
425.3 |
47.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.013 ± 0.535 | 2.251 ± 0.329 |
5.039 ± 0.759 | 6.214 ± 0.798 |
6.449 ± 1.109 | 5.778 ± 0.419 |
2.754 ± 0.596 | 6.349 ± 0.695 |
7.054 ± 1.008 | 10.648 ± 1.916 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.78 ± 0.298 | 4.232 ± 0.565 |
3.93 ± 0.566 | 2.788 ± 0.448 |
5.173 ± 0.524 | 9.271 ± 0.577 |
4.636 ± 1.056 | 6.013 ± 0.684 |
0.773 ± 0.157 | 2.855 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
