
Halomarina oriensis
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halomarina
Average proteome isoelectric point is 5.09
Get precalculated fractions of proteins
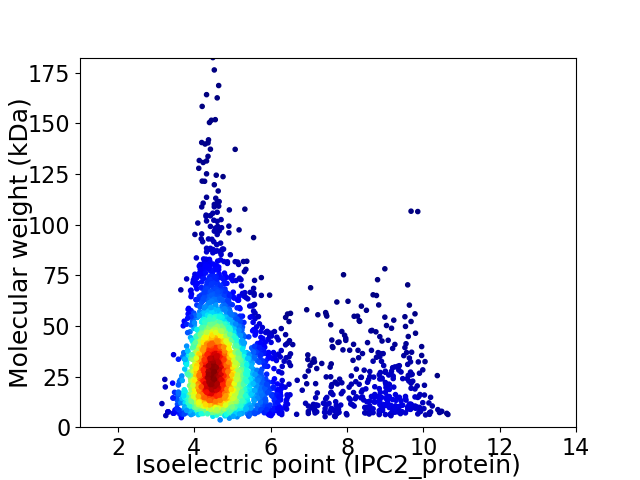
Virtual 2D-PAGE plot for 3340 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3P3RE93|A0A3P3RE93_9EURY Right-handed parallel beta-helix repeat-containing protein OS=Halomarina oriensis OX=671145 GN=EIK79_06035 PE=4 SV=1
MM1 pKa = 7.04TQDD4 pKa = 3.08NHH6 pKa = 5.91NTDD9 pKa = 3.37TTDD12 pKa = 3.86DD13 pKa = 3.83RR14 pKa = 11.84STDD17 pKa = 3.34GTRR20 pKa = 11.84SSWHH24 pKa = 6.09RR25 pKa = 11.84LSGVFEE31 pKa = 4.61DD32 pKa = 3.6SSRR35 pKa = 11.84RR36 pKa = 11.84EE37 pKa = 3.85FLRR40 pKa = 11.84AGAAIGAGGLALGLGNTATAAKK62 pKa = 10.38VEE64 pKa = 4.69EE65 pKa = 4.7ICNSDD70 pKa = 3.46YY71 pKa = 10.94GTIDD75 pKa = 3.25VADD78 pKa = 4.78GFTLMDD84 pKa = 4.14NRR86 pKa = 11.84WGNQNAHH93 pKa = 4.2QCIWYY98 pKa = 9.9NDD100 pKa = 3.96DD101 pKa = 3.3GSYY104 pKa = 11.06GYY106 pKa = 10.91DD107 pKa = 3.18FDD109 pKa = 5.73ASNTGGGINYY119 pKa = 9.21PEE121 pKa = 4.28VFIGTRR127 pKa = 11.84PWGASTGVVEE137 pKa = 6.68FPIQRR142 pKa = 11.84HH143 pKa = 5.52DD144 pKa = 3.34VDD146 pKa = 4.0EE147 pKa = 4.79FVMEE151 pKa = 4.3VDD153 pKa = 3.65ADD155 pKa = 4.12LDD157 pKa = 3.66ISGGEE162 pKa = 3.95WDD164 pKa = 4.08WAEE167 pKa = 4.21EE168 pKa = 3.91WWLMEE173 pKa = 4.74QPPTQQPEE181 pKa = 4.02THH183 pKa = 6.15QYY185 pKa = 11.07EE186 pKa = 4.69VMLLLDD192 pKa = 3.28WGGGHH197 pKa = 7.03DD198 pKa = 4.39HH199 pKa = 7.06GAVQDD204 pKa = 3.99PSAWTDD210 pKa = 3.08RR211 pKa = 11.84FGNTIDD217 pKa = 3.84LWTTYY222 pKa = 11.25DD223 pKa = 3.62SGGTDD228 pKa = 2.76AYY230 pKa = 10.75FYY232 pKa = 10.59IFRR235 pKa = 11.84IQGGHH240 pKa = 7.13DD241 pKa = 3.18GGKK244 pKa = 9.22IDD246 pKa = 3.45MAEE249 pKa = 3.67IVEE252 pKa = 4.24YY253 pKa = 10.65LSSNEE258 pKa = 3.83GVRR261 pKa = 11.84GDD263 pKa = 3.5LWLSGIEE270 pKa = 4.61LGNEE274 pKa = 3.66YY275 pKa = 9.66WAGAVGEE282 pKa = 4.42TTYY285 pKa = 10.22NTFNVTINGSTYY297 pKa = 10.55TSGTGDD303 pKa = 3.5GGSPPTDD310 pKa = 3.41TPTDD314 pKa = 3.78TPTDD318 pKa = 3.68TPGDD322 pKa = 3.57GLVINDD328 pKa = 3.83YY329 pKa = 11.26DD330 pKa = 4.87GDD332 pKa = 4.35PSWSSNTNDD341 pKa = 3.13LGQWCGAGSFANGSGTVSNGALVLEE366 pKa = 4.36YY367 pKa = 10.98DD368 pKa = 3.15NGGWYY373 pKa = 9.47QEE375 pKa = 4.04QLNQSVTDD383 pKa = 3.47YY384 pKa = 9.62STLVLRR390 pKa = 11.84VSGASGGEE398 pKa = 3.83EE399 pKa = 3.61SDD401 pKa = 3.25ITVEE405 pKa = 4.01MGGVSAPLADD415 pKa = 3.67VTSDD419 pKa = 4.02SIATNMSDD427 pKa = 4.38VAIDD431 pKa = 3.79LQAAGVDD438 pKa = 3.76RR439 pKa = 11.84SSSSLSLRR447 pKa = 11.84LNFWGGGSSTLQIEE461 pKa = 4.76EE462 pKa = 4.02IALVV466 pKa = 3.47
MM1 pKa = 7.04TQDD4 pKa = 3.08NHH6 pKa = 5.91NTDD9 pKa = 3.37TTDD12 pKa = 3.86DD13 pKa = 3.83RR14 pKa = 11.84STDD17 pKa = 3.34GTRR20 pKa = 11.84SSWHH24 pKa = 6.09RR25 pKa = 11.84LSGVFEE31 pKa = 4.61DD32 pKa = 3.6SSRR35 pKa = 11.84RR36 pKa = 11.84EE37 pKa = 3.85FLRR40 pKa = 11.84AGAAIGAGGLALGLGNTATAAKK62 pKa = 10.38VEE64 pKa = 4.69EE65 pKa = 4.7ICNSDD70 pKa = 3.46YY71 pKa = 10.94GTIDD75 pKa = 3.25VADD78 pKa = 4.78GFTLMDD84 pKa = 4.14NRR86 pKa = 11.84WGNQNAHH93 pKa = 4.2QCIWYY98 pKa = 9.9NDD100 pKa = 3.96DD101 pKa = 3.3GSYY104 pKa = 11.06GYY106 pKa = 10.91DD107 pKa = 3.18FDD109 pKa = 5.73ASNTGGGINYY119 pKa = 9.21PEE121 pKa = 4.28VFIGTRR127 pKa = 11.84PWGASTGVVEE137 pKa = 6.68FPIQRR142 pKa = 11.84HH143 pKa = 5.52DD144 pKa = 3.34VDD146 pKa = 4.0EE147 pKa = 4.79FVMEE151 pKa = 4.3VDD153 pKa = 3.65ADD155 pKa = 4.12LDD157 pKa = 3.66ISGGEE162 pKa = 3.95WDD164 pKa = 4.08WAEE167 pKa = 4.21EE168 pKa = 3.91WWLMEE173 pKa = 4.74QPPTQQPEE181 pKa = 4.02THH183 pKa = 6.15QYY185 pKa = 11.07EE186 pKa = 4.69VMLLLDD192 pKa = 3.28WGGGHH197 pKa = 7.03DD198 pKa = 4.39HH199 pKa = 7.06GAVQDD204 pKa = 3.99PSAWTDD210 pKa = 3.08RR211 pKa = 11.84FGNTIDD217 pKa = 3.84LWTTYY222 pKa = 11.25DD223 pKa = 3.62SGGTDD228 pKa = 2.76AYY230 pKa = 10.75FYY232 pKa = 10.59IFRR235 pKa = 11.84IQGGHH240 pKa = 7.13DD241 pKa = 3.18GGKK244 pKa = 9.22IDD246 pKa = 3.45MAEE249 pKa = 3.67IVEE252 pKa = 4.24YY253 pKa = 10.65LSSNEE258 pKa = 3.83GVRR261 pKa = 11.84GDD263 pKa = 3.5LWLSGIEE270 pKa = 4.61LGNEE274 pKa = 3.66YY275 pKa = 9.66WAGAVGEE282 pKa = 4.42TTYY285 pKa = 10.22NTFNVTINGSTYY297 pKa = 10.55TSGTGDD303 pKa = 3.5GGSPPTDD310 pKa = 3.41TPTDD314 pKa = 3.78TPTDD318 pKa = 3.68TPGDD322 pKa = 3.57GLVINDD328 pKa = 3.83YY329 pKa = 11.26DD330 pKa = 4.87GDD332 pKa = 4.35PSWSSNTNDD341 pKa = 3.13LGQWCGAGSFANGSGTVSNGALVLEE366 pKa = 4.36YY367 pKa = 10.98DD368 pKa = 3.15NGGWYY373 pKa = 9.47QEE375 pKa = 4.04QLNQSVTDD383 pKa = 3.47YY384 pKa = 9.62STLVLRR390 pKa = 11.84VSGASGGEE398 pKa = 3.83EE399 pKa = 3.61SDD401 pKa = 3.25ITVEE405 pKa = 4.01MGGVSAPLADD415 pKa = 3.67VTSDD419 pKa = 4.02SIATNMSDD427 pKa = 4.38VAIDD431 pKa = 3.79LQAAGVDD438 pKa = 3.76RR439 pKa = 11.84SSSSLSLRR447 pKa = 11.84LNFWGGGSSTLQIEE461 pKa = 4.76EE462 pKa = 4.02IALVV466 pKa = 3.47
Molecular weight: 50.04 kDa
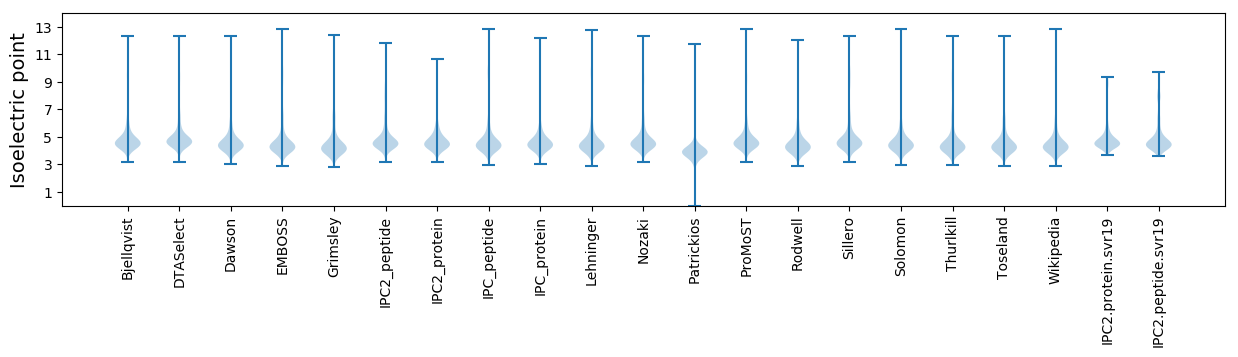
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P3RJY3|A0A3P3RJY3_9EURY GYD domain-containing protein OS=Halomarina oriensis OX=671145 GN=EIK79_03640 PE=4 SV=1
MM1 pKa = 7.56SEE3 pKa = 4.35RR4 pKa = 11.84EE5 pKa = 4.13SSVTRR10 pKa = 11.84PRR12 pKa = 11.84SDD14 pKa = 3.2SKK16 pKa = 10.63PLQWLFDD23 pKa = 4.18PSLLALCGVSVLLVAVGGLVQVSFGSYY50 pKa = 8.86QMSLTEE56 pKa = 3.57VWQAVFNPAVLFDD69 pKa = 4.66LRR71 pKa = 11.84WLSKK75 pKa = 10.61FILGDD80 pKa = 3.41SVMRR84 pKa = 11.84SITGFTGTLPEE95 pKa = 5.5LDD97 pKa = 3.45TSTTVVWNIRR107 pKa = 11.84LPRR110 pKa = 11.84VLLAVFVGMNLAVSGAIFQAVTRR133 pKa = 11.84NEE135 pKa = 3.94LASPFVLGISHH146 pKa = 7.18GAGLVILLTLVVFTGLTAWLPIVAALGGAVAFTIVYY182 pKa = 10.05AIAWKK187 pKa = 10.63NGTSPVRR194 pKa = 11.84LVLAGIIVATVFQSLQRR211 pKa = 11.84SLFFFATDD219 pKa = 2.64IGTAQTAMAWMTGSLTGVGWTQFRR243 pKa = 11.84IALPWTILVMALSIISARR261 pKa = 11.84QLNVLLLGQRR271 pKa = 11.84TAKK274 pKa = 10.1SLGMSVEE281 pKa = 3.99RR282 pKa = 11.84VRR284 pKa = 11.84FALSGVAVLAAAVSIAVAGVVSFVGLIVPHH314 pKa = 6.29LVRR317 pKa = 11.84TLVGNDD323 pKa = 3.21HH324 pKa = 6.84KK325 pKa = 11.28KK326 pKa = 11.02LIVGCLFTGPALMVVADD343 pKa = 3.74VGARR347 pKa = 11.84LGPSILIGAANQQLPVGVVTGLIGGPYY374 pKa = 9.99FLYY377 pKa = 10.82LMRR380 pKa = 11.84KK381 pKa = 8.13RR382 pKa = 11.84QQLGDD387 pKa = 3.51VV388 pKa = 3.84
MM1 pKa = 7.56SEE3 pKa = 4.35RR4 pKa = 11.84EE5 pKa = 4.13SSVTRR10 pKa = 11.84PRR12 pKa = 11.84SDD14 pKa = 3.2SKK16 pKa = 10.63PLQWLFDD23 pKa = 4.18PSLLALCGVSVLLVAVGGLVQVSFGSYY50 pKa = 8.86QMSLTEE56 pKa = 3.57VWQAVFNPAVLFDD69 pKa = 4.66LRR71 pKa = 11.84WLSKK75 pKa = 10.61FILGDD80 pKa = 3.41SVMRR84 pKa = 11.84SITGFTGTLPEE95 pKa = 5.5LDD97 pKa = 3.45TSTTVVWNIRR107 pKa = 11.84LPRR110 pKa = 11.84VLLAVFVGMNLAVSGAIFQAVTRR133 pKa = 11.84NEE135 pKa = 3.94LASPFVLGISHH146 pKa = 7.18GAGLVILLTLVVFTGLTAWLPIVAALGGAVAFTIVYY182 pKa = 10.05AIAWKK187 pKa = 10.63NGTSPVRR194 pKa = 11.84LVLAGIIVATVFQSLQRR211 pKa = 11.84SLFFFATDD219 pKa = 2.64IGTAQTAMAWMTGSLTGVGWTQFRR243 pKa = 11.84IALPWTILVMALSIISARR261 pKa = 11.84QLNVLLLGQRR271 pKa = 11.84TAKK274 pKa = 10.1SLGMSVEE281 pKa = 3.99RR282 pKa = 11.84VRR284 pKa = 11.84FALSGVAVLAAAVSIAVAGVVSFVGLIVPHH314 pKa = 6.29LVRR317 pKa = 11.84TLVGNDD323 pKa = 3.21HH324 pKa = 6.84KK325 pKa = 11.28KK326 pKa = 11.02LIVGCLFTGPALMVVADD343 pKa = 3.74VGARR347 pKa = 11.84LGPSILIGAANQQLPVGVVTGLIGGPYY374 pKa = 9.99FLYY377 pKa = 10.82LMRR380 pKa = 11.84KK381 pKa = 8.13RR382 pKa = 11.84QQLGDD387 pKa = 3.51VV388 pKa = 3.84
Molecular weight: 41.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
990830 |
32 |
1653 |
296.7 |
32.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.49 ± 0.047 | 0.846 ± 0.015 |
7.56 ± 0.045 | 7.822 ± 0.047 |
3.355 ± 0.029 | 7.917 ± 0.038 |
2.313 ± 0.021 | 5.064 ± 0.033 |
2.125 ± 0.021 | 8.864 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.818 ± 0.016 | 2.843 ± 0.027 |
4.649 ± 0.024 | 3.116 ± 0.025 |
6.387 ± 0.038 | 6.09 ± 0.032 |
7.338 ± 0.038 | 8.324 ± 0.038 |
1.204 ± 0.017 | 2.876 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
