
Lake Sarah-associated circular virus-9
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.15
Get precalculated fractions of proteins
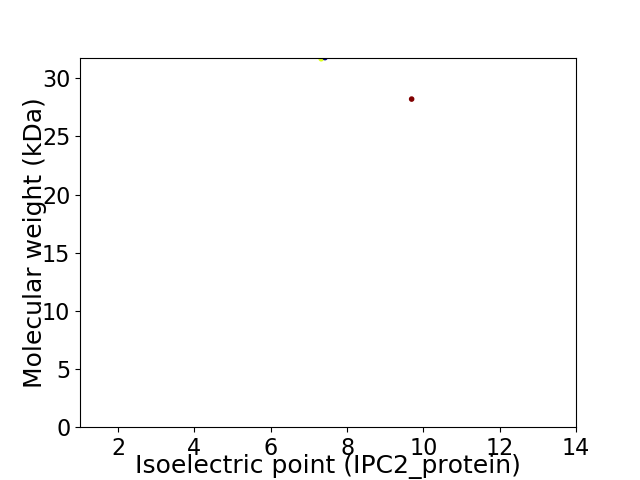
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A126GA58|A0A126GA58_9VIRU Coat protein OS=Lake Sarah-associated circular virus-9 OX=1685786 PE=4 SV=1
MM1 pKa = 7.36SRR3 pKa = 11.84QLRR6 pKa = 11.84HH7 pKa = 6.04FCFTLNNYY15 pKa = 8.59VEE17 pKa = 4.67EE18 pKa = 4.05EE19 pKa = 4.62DD20 pKa = 3.65IPRR23 pKa = 11.84LQGFFEE29 pKa = 4.22QEE31 pKa = 3.16AKK33 pKa = 10.05YY34 pKa = 9.55WIIGRR39 pKa = 11.84EE40 pKa = 4.2VGDD43 pKa = 3.42SGTPHH48 pKa = 6.05LQGYY52 pKa = 10.45ASLQRR57 pKa = 11.84RR58 pKa = 11.84HH59 pKa = 5.44TFNVVRR65 pKa = 11.84DD66 pKa = 3.95KK67 pKa = 11.38LGSRR71 pKa = 11.84CHH73 pKa = 7.04IEE75 pKa = 3.96GARR78 pKa = 11.84GSARR82 pKa = 11.84QNRR85 pKa = 11.84EE86 pKa = 3.7YY87 pKa = 10.65CSKK90 pKa = 10.22GGNYY94 pKa = 9.3IEE96 pKa = 4.91GGSINEE102 pKa = 4.23GAASGKK108 pKa = 10.3SRR110 pKa = 11.84DD111 pKa = 3.56EE112 pKa = 4.19LGRR115 pKa = 11.84SFMAAVEE122 pKa = 4.31LGNSGVVEE130 pKa = 4.19YY131 pKa = 11.22ANSFPGAYY139 pKa = 8.97IFSGSNMLRR148 pKa = 11.84NALALKK154 pKa = 10.19PPIEE158 pKa = 4.29RR159 pKa = 11.84PNIKK163 pKa = 9.63VQWIYY168 pKa = 11.46GEE170 pKa = 4.19PGVGKK175 pKa = 10.39SRR177 pKa = 11.84LAHH180 pKa = 6.04SSLPEE185 pKa = 4.14AYY187 pKa = 10.18VKK189 pKa = 10.49EE190 pKa = 4.48PRR192 pKa = 11.84TKK194 pKa = 8.11WWNGYY199 pKa = 7.28MCEE202 pKa = 4.39KK203 pKa = 10.7NVIIDD208 pKa = 3.67DD209 pKa = 4.53FGPNGIDD216 pKa = 3.32INHH219 pKa = 7.46LLRR222 pKa = 11.84WFDD225 pKa = 3.45RR226 pKa = 11.84YY227 pKa = 10.77KK228 pKa = 11.0CLVEE232 pKa = 4.4NKK234 pKa = 10.12GGMIALYY241 pKa = 10.2ADD243 pKa = 3.44NFIITSNFHH252 pKa = 6.26PDD254 pKa = 3.09EE255 pKa = 4.51VFSFAGVPNVQLPALMRR272 pKa = 11.84RR273 pKa = 11.84IEE275 pKa = 4.18LVHH278 pKa = 6.1MNN280 pKa = 3.9
MM1 pKa = 7.36SRR3 pKa = 11.84QLRR6 pKa = 11.84HH7 pKa = 6.04FCFTLNNYY15 pKa = 8.59VEE17 pKa = 4.67EE18 pKa = 4.05EE19 pKa = 4.62DD20 pKa = 3.65IPRR23 pKa = 11.84LQGFFEE29 pKa = 4.22QEE31 pKa = 3.16AKK33 pKa = 10.05YY34 pKa = 9.55WIIGRR39 pKa = 11.84EE40 pKa = 4.2VGDD43 pKa = 3.42SGTPHH48 pKa = 6.05LQGYY52 pKa = 10.45ASLQRR57 pKa = 11.84RR58 pKa = 11.84HH59 pKa = 5.44TFNVVRR65 pKa = 11.84DD66 pKa = 3.95KK67 pKa = 11.38LGSRR71 pKa = 11.84CHH73 pKa = 7.04IEE75 pKa = 3.96GARR78 pKa = 11.84GSARR82 pKa = 11.84QNRR85 pKa = 11.84EE86 pKa = 3.7YY87 pKa = 10.65CSKK90 pKa = 10.22GGNYY94 pKa = 9.3IEE96 pKa = 4.91GGSINEE102 pKa = 4.23GAASGKK108 pKa = 10.3SRR110 pKa = 11.84DD111 pKa = 3.56EE112 pKa = 4.19LGRR115 pKa = 11.84SFMAAVEE122 pKa = 4.31LGNSGVVEE130 pKa = 4.19YY131 pKa = 11.22ANSFPGAYY139 pKa = 8.97IFSGSNMLRR148 pKa = 11.84NALALKK154 pKa = 10.19PPIEE158 pKa = 4.29RR159 pKa = 11.84PNIKK163 pKa = 9.63VQWIYY168 pKa = 11.46GEE170 pKa = 4.19PGVGKK175 pKa = 10.39SRR177 pKa = 11.84LAHH180 pKa = 6.04SSLPEE185 pKa = 4.14AYY187 pKa = 10.18VKK189 pKa = 10.49EE190 pKa = 4.48PRR192 pKa = 11.84TKK194 pKa = 8.11WWNGYY199 pKa = 7.28MCEE202 pKa = 4.39KK203 pKa = 10.7NVIIDD208 pKa = 3.67DD209 pKa = 4.53FGPNGIDD216 pKa = 3.32INHH219 pKa = 7.46LLRR222 pKa = 11.84WFDD225 pKa = 3.45RR226 pKa = 11.84YY227 pKa = 10.77KK228 pKa = 11.0CLVEE232 pKa = 4.4NKK234 pKa = 10.12GGMIALYY241 pKa = 10.2ADD243 pKa = 3.44NFIITSNFHH252 pKa = 6.26PDD254 pKa = 3.09EE255 pKa = 4.51VFSFAGVPNVQLPALMRR272 pKa = 11.84RR273 pKa = 11.84IEE275 pKa = 4.18LVHH278 pKa = 6.1MNN280 pKa = 3.9
Molecular weight: 31.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA58|A0A126GA58_9VIRU Coat protein OS=Lake Sarah-associated circular virus-9 OX=1685786 PE=4 SV=1
MM1 pKa = 7.45ARR3 pKa = 11.84YY4 pKa = 7.72NQKK7 pKa = 9.97RR8 pKa = 11.84KK9 pKa = 9.77RR10 pKa = 11.84SSGPFRR16 pKa = 11.84SFKK19 pKa = 10.19KK20 pKa = 10.17RR21 pKa = 11.84RR22 pKa = 11.84FARR25 pKa = 11.84KK26 pKa = 8.93KK27 pKa = 10.64ARR29 pKa = 11.84TSYY32 pKa = 10.26INSVNGRR39 pKa = 11.84VTSTPFFMGRR49 pKa = 11.84KK50 pKa = 7.6MRR52 pKa = 11.84PKK54 pKa = 10.17KK55 pKa = 10.35YY56 pKa = 10.54RR57 pKa = 11.84NILWNATMGSEE68 pKa = 5.16KK69 pKa = 10.99YY70 pKa = 10.36NLNNTIALTVSTPASLVNYY89 pKa = 8.38TYY91 pKa = 11.29VNQQMFVGLDD101 pKa = 3.96DD102 pKa = 3.27ITKK105 pKa = 8.22YY106 pKa = 9.66TNSNGPNPTATLGKK120 pKa = 7.96VTMRR124 pKa = 11.84GGYY127 pKa = 9.89SKK129 pKa = 10.23FTLSSKK135 pKa = 10.64DD136 pKa = 3.74DD137 pKa = 3.53EE138 pKa = 5.01VIDD141 pKa = 3.74YY142 pKa = 10.19RR143 pKa = 11.84LYY145 pKa = 10.97AIWVKK150 pKa = 10.7PNGVPPGAGALGKK163 pKa = 10.69SIDD166 pKa = 3.84IAHH169 pKa = 6.76SATDD173 pKa = 3.42TSNEE177 pKa = 3.88SVRR180 pKa = 11.84ILKK183 pKa = 9.17QWSGLLDD190 pKa = 4.0RR191 pKa = 11.84SQSASFFFRR200 pKa = 11.84AKK202 pKa = 9.95IKK204 pKa = 10.77QYY206 pKa = 9.6DD207 pKa = 3.67TTMFTAGTDD216 pKa = 3.29CMYY219 pKa = 10.42WYY221 pKa = 10.36LGVGNTIDD229 pKa = 3.36ATAVSVSYY237 pKa = 10.32IQSFNCSMTMDD248 pKa = 3.73VTAA251 pKa = 4.94
MM1 pKa = 7.45ARR3 pKa = 11.84YY4 pKa = 7.72NQKK7 pKa = 9.97RR8 pKa = 11.84KK9 pKa = 9.77RR10 pKa = 11.84SSGPFRR16 pKa = 11.84SFKK19 pKa = 10.19KK20 pKa = 10.17RR21 pKa = 11.84RR22 pKa = 11.84FARR25 pKa = 11.84KK26 pKa = 8.93KK27 pKa = 10.64ARR29 pKa = 11.84TSYY32 pKa = 10.26INSVNGRR39 pKa = 11.84VTSTPFFMGRR49 pKa = 11.84KK50 pKa = 7.6MRR52 pKa = 11.84PKK54 pKa = 10.17KK55 pKa = 10.35YY56 pKa = 10.54RR57 pKa = 11.84NILWNATMGSEE68 pKa = 5.16KK69 pKa = 10.99YY70 pKa = 10.36NLNNTIALTVSTPASLVNYY89 pKa = 8.38TYY91 pKa = 11.29VNQQMFVGLDD101 pKa = 3.96DD102 pKa = 3.27ITKK105 pKa = 8.22YY106 pKa = 9.66TNSNGPNPTATLGKK120 pKa = 7.96VTMRR124 pKa = 11.84GGYY127 pKa = 9.89SKK129 pKa = 10.23FTLSSKK135 pKa = 10.64DD136 pKa = 3.74DD137 pKa = 3.53EE138 pKa = 5.01VIDD141 pKa = 3.74YY142 pKa = 10.19RR143 pKa = 11.84LYY145 pKa = 10.97AIWVKK150 pKa = 10.7PNGVPPGAGALGKK163 pKa = 10.69SIDD166 pKa = 3.84IAHH169 pKa = 6.76SATDD173 pKa = 3.42TSNEE177 pKa = 3.88SVRR180 pKa = 11.84ILKK183 pKa = 9.17QWSGLLDD190 pKa = 4.0RR191 pKa = 11.84SQSASFFFRR200 pKa = 11.84AKK202 pKa = 9.95IKK204 pKa = 10.77QYY206 pKa = 9.6DD207 pKa = 3.67TTMFTAGTDD216 pKa = 3.29CMYY219 pKa = 10.42WYY221 pKa = 10.36LGVGNTIDD229 pKa = 3.36ATAVSVSYY237 pKa = 10.32IQSFNCSMTMDD248 pKa = 3.73VTAA251 pKa = 4.94
Molecular weight: 28.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
531 |
251 |
280 |
265.5 |
29.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.78 ± 0.247 | 1.318 ± 0.329 |
4.143 ± 0.402 | 4.52 ± 2.095 |
4.896 ± 0.073 | 8.475 ± 1.072 |
1.695 ± 0.817 | 5.65 ± 0.547 |
5.838 ± 1.091 | 6.215 ± 0.652 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.202 ± 0.493 | 6.968 ± 0.123 |
4.143 ± 0.351 | 2.825 ± 0.023 |
7.156 ± 0.242 | 8.286 ± 1.055 |
5.461 ± 2.584 | 5.838 ± 0.087 |
1.695 ± 0.064 | 4.896 ± 0.429 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
