
Heliothis virescens ascovirus 3e (HvAV-3e)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Ascoviridae; Ascovirus; Heliothis virescens ascovirus 3a
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
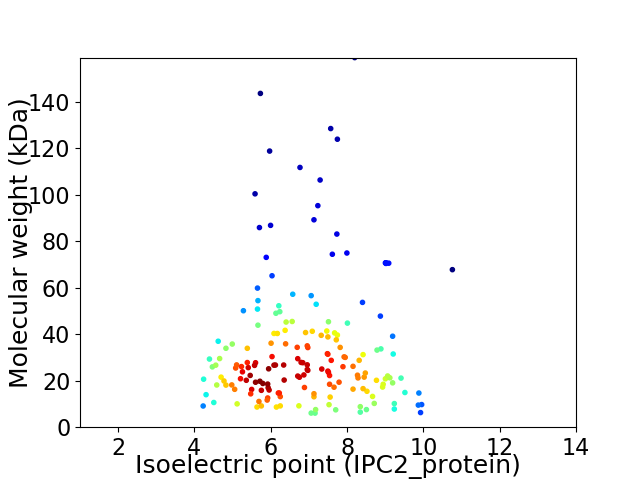
Virtual 2D-PAGE plot for 178 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4KXF0|A4KXF0_HVAVE NAD-glutamate dehydrogenase OS=Heliothis virescens ascovirus 3e OX=260797 PE=4 SV=1
MM1 pKa = 7.47NEE3 pKa = 3.51AEE5 pKa = 5.13LRR7 pKa = 11.84VQYY10 pKa = 10.42EE11 pKa = 4.27SEE13 pKa = 4.51DD14 pKa = 4.03EE15 pKa = 4.52ITLEE19 pKa = 3.87WLKK22 pKa = 9.85TLGVNDD28 pKa = 4.23ADD30 pKa = 3.62EE31 pKa = 5.47CVAQCHH37 pKa = 5.32VSHH40 pKa = 6.38EE41 pKa = 4.14QALARR46 pKa = 11.84IKK48 pKa = 10.67RR49 pKa = 11.84AVDD52 pKa = 3.18EE53 pKa = 5.46CDD55 pKa = 3.52DD56 pKa = 3.63QSVIDD61 pKa = 4.2NEE63 pKa = 4.49VNRR66 pKa = 11.84PPSADD71 pKa = 3.16NVSYY75 pKa = 8.99TAEE78 pKa = 3.96EE79 pKa = 4.68VYY81 pKa = 10.86DD82 pKa = 4.01AVSLIRR88 pKa = 11.84DD89 pKa = 3.8RR90 pKa = 11.84GRR92 pKa = 11.84MVNITNNYY100 pKa = 9.1NFFGSPPRR108 pKa = 11.84PSLVPPPPPPPPPYY122 pKa = 10.0EE123 pKa = 3.82QQ124 pKa = 3.68
MM1 pKa = 7.47NEE3 pKa = 3.51AEE5 pKa = 5.13LRR7 pKa = 11.84VQYY10 pKa = 10.42EE11 pKa = 4.27SEE13 pKa = 4.51DD14 pKa = 4.03EE15 pKa = 4.52ITLEE19 pKa = 3.87WLKK22 pKa = 9.85TLGVNDD28 pKa = 4.23ADD30 pKa = 3.62EE31 pKa = 5.47CVAQCHH37 pKa = 5.32VSHH40 pKa = 6.38EE41 pKa = 4.14QALARR46 pKa = 11.84IKK48 pKa = 10.67RR49 pKa = 11.84AVDD52 pKa = 3.18EE53 pKa = 5.46CDD55 pKa = 3.52DD56 pKa = 3.63QSVIDD61 pKa = 4.2NEE63 pKa = 4.49VNRR66 pKa = 11.84PPSADD71 pKa = 3.16NVSYY75 pKa = 8.99TAEE78 pKa = 3.96EE79 pKa = 4.68VYY81 pKa = 10.86DD82 pKa = 4.01AVSLIRR88 pKa = 11.84DD89 pKa = 3.8RR90 pKa = 11.84GRR92 pKa = 11.84MVNITNNYY100 pKa = 9.1NFFGSPPRR108 pKa = 11.84PSLVPPPPPPPPPYY122 pKa = 10.0EE123 pKa = 3.82QQ124 pKa = 3.68
Molecular weight: 13.98 kDa
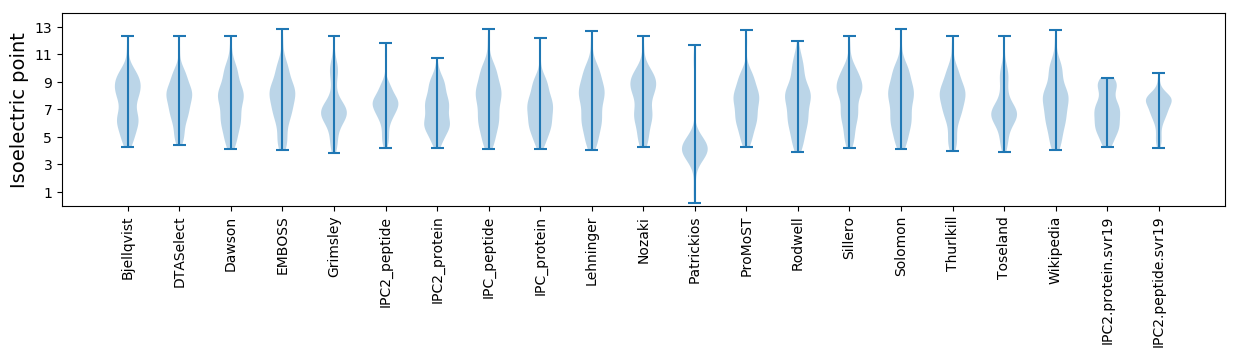
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4KXB7|A4KXB7_HVAVE Putative nucleic acid-binding protein OS=Heliothis virescens ascovirus 3e OX=260797 PE=4 SV=1
MM1 pKa = 7.73ASRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.66PSRR9 pKa = 11.84LTAAQCEE16 pKa = 4.29TFIRR20 pKa = 11.84NNKK23 pKa = 8.58AVSPLTNKK31 pKa = 10.24PIDD34 pKa = 3.75VYY36 pKa = 11.32GRR38 pKa = 11.84AAARR42 pKa = 11.84FRR44 pKa = 11.84RR45 pKa = 11.84DD46 pKa = 3.7CNLSPPPTKK55 pKa = 9.91YY56 pKa = 9.46TSTVCKK62 pKa = 10.3KK63 pKa = 9.77FLANKK68 pKa = 9.71SVSPYY73 pKa = 9.63SGRR76 pKa = 11.84PIKK79 pKa = 10.58RR80 pKa = 11.84GAKK83 pKa = 9.77LYY85 pKa = 11.04NDD87 pKa = 3.81LTKK90 pKa = 10.35HH91 pKa = 5.73CSGTRR96 pKa = 11.84SSSPARR102 pKa = 11.84SPARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84VIRR112 pKa = 11.84SPSPNRR118 pKa = 11.84RR119 pKa = 11.84SSSPRR124 pKa = 11.84RR125 pKa = 11.84SASPQRR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84ASPQRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84ASPQRR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84ASPDD151 pKa = 2.58RR152 pKa = 11.84SKK154 pKa = 10.64PAKK157 pKa = 8.89RR158 pKa = 11.84TAANADD164 pKa = 3.61TRR166 pKa = 11.84PDD168 pKa = 3.15LCATFSRR175 pKa = 11.84NEE177 pKa = 4.28SINPITGKK185 pKa = 10.6KK186 pKa = 10.32LIGTSPIRR194 pKa = 11.84KK195 pKa = 8.42AWHH198 pKa = 6.02RR199 pKa = 11.84MCAGTPNTRR208 pKa = 11.84ATKK211 pKa = 10.57CMAFDD216 pKa = 4.94KK217 pKa = 10.78NDD219 pKa = 3.44KK220 pKa = 10.4KK221 pKa = 11.18NPFTGRR227 pKa = 11.84SIRR230 pKa = 11.84PEE232 pKa = 3.65QPAYY236 pKa = 10.69RR237 pKa = 11.84MVYY240 pKa = 9.71SMCHH244 pKa = 4.51GVPYY248 pKa = 10.35RR249 pKa = 11.84SPKK252 pKa = 7.71RR253 pKa = 11.84TRR255 pKa = 11.84RR256 pKa = 11.84SPVRR260 pKa = 11.84RR261 pKa = 11.84SPSPRR266 pKa = 11.84PYY268 pKa = 9.42TATSVTRR275 pKa = 11.84KK276 pKa = 8.58YY277 pKa = 10.96RR278 pKa = 11.84RR279 pKa = 11.84IKK281 pKa = 8.95TPARR285 pKa = 11.84SRR287 pKa = 11.84SRR289 pKa = 11.84SRR291 pKa = 11.84SNSVGRR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84TTAVKK304 pKa = 10.56SRR306 pKa = 11.84TKK308 pKa = 10.71SPARR312 pKa = 11.84RR313 pKa = 11.84QSVARR318 pKa = 11.84SRR320 pKa = 11.84SRR322 pKa = 11.84SKK324 pKa = 10.99SPVRR328 pKa = 11.84KK329 pKa = 5.63TTRR332 pKa = 11.84SRR334 pKa = 11.84TKK336 pKa = 10.25SPARR340 pKa = 11.84RR341 pKa = 11.84QSVARR346 pKa = 11.84SRR348 pKa = 11.84SRR350 pKa = 11.84TKK352 pKa = 10.66SPARR356 pKa = 11.84RR357 pKa = 11.84QSVARR362 pKa = 11.84SRR364 pKa = 11.84SRR366 pKa = 11.84SKK368 pKa = 10.93SPARR372 pKa = 11.84RR373 pKa = 11.84QSVARR378 pKa = 11.84SRR380 pKa = 11.84SRR382 pKa = 11.84SKK384 pKa = 10.93SPARR388 pKa = 11.84RR389 pKa = 11.84QSVAKK394 pKa = 10.05SRR396 pKa = 11.84SRR398 pKa = 11.84SKK400 pKa = 10.95SPARR404 pKa = 11.84RR405 pKa = 11.84QSVAMSRR412 pKa = 11.84SRR414 pKa = 11.84SRR416 pKa = 11.84SRR418 pKa = 11.84SRR420 pKa = 11.84QPMTAMRR427 pKa = 11.84RR428 pKa = 11.84STSRR432 pKa = 11.84ARR434 pKa = 11.84SRR436 pKa = 11.84SKK438 pKa = 10.59SRR440 pKa = 11.84SRR442 pKa = 11.84KK443 pKa = 9.7AMTASRR449 pKa = 11.84SRR451 pKa = 11.84SRR453 pKa = 11.84SVSRR457 pKa = 11.84YY458 pKa = 7.72MINPNFVPTTTKK470 pKa = 10.14KK471 pKa = 10.55KK472 pKa = 9.82RR473 pKa = 11.84VSPVSRR479 pKa = 11.84GRR481 pKa = 11.84INNSRR486 pKa = 11.84SRR488 pKa = 11.84SASRR492 pKa = 11.84ARR494 pKa = 11.84SGGLSPYY501 pKa = 9.82RR502 pKa = 11.84GRR504 pKa = 11.84VLLSPIPDD512 pKa = 3.73GATPMSRR519 pKa = 11.84SQLINIANNMNIAEE533 pKa = 4.24LRR535 pKa = 11.84HH536 pKa = 5.35IVVSNGFQPVRR547 pKa = 11.84VAQNTTQSQLLNIVKK562 pKa = 9.21FQIRR566 pKa = 11.84EE567 pKa = 4.25GNLKK571 pKa = 9.24WLPRR575 pKa = 11.84NDD577 pKa = 3.38QDD579 pKa = 3.62VPTYY583 pKa = 8.27YY584 pKa = 10.27TSSRR588 pKa = 11.84PFADD592 pKa = 3.04RR593 pKa = 11.84MKK595 pKa = 11.04KK596 pKa = 9.43NN597 pKa = 3.25
MM1 pKa = 7.73ASRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.66PSRR9 pKa = 11.84LTAAQCEE16 pKa = 4.29TFIRR20 pKa = 11.84NNKK23 pKa = 8.58AVSPLTNKK31 pKa = 10.24PIDD34 pKa = 3.75VYY36 pKa = 11.32GRR38 pKa = 11.84AAARR42 pKa = 11.84FRR44 pKa = 11.84RR45 pKa = 11.84DD46 pKa = 3.7CNLSPPPTKK55 pKa = 9.91YY56 pKa = 9.46TSTVCKK62 pKa = 10.3KK63 pKa = 9.77FLANKK68 pKa = 9.71SVSPYY73 pKa = 9.63SGRR76 pKa = 11.84PIKK79 pKa = 10.58RR80 pKa = 11.84GAKK83 pKa = 9.77LYY85 pKa = 11.04NDD87 pKa = 3.81LTKK90 pKa = 10.35HH91 pKa = 5.73CSGTRR96 pKa = 11.84SSSPARR102 pKa = 11.84SPARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84VIRR112 pKa = 11.84SPSPNRR118 pKa = 11.84RR119 pKa = 11.84SSSPRR124 pKa = 11.84RR125 pKa = 11.84SASPQRR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84ASPQRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84ASPQRR145 pKa = 11.84RR146 pKa = 11.84RR147 pKa = 11.84ASPDD151 pKa = 2.58RR152 pKa = 11.84SKK154 pKa = 10.64PAKK157 pKa = 8.89RR158 pKa = 11.84TAANADD164 pKa = 3.61TRR166 pKa = 11.84PDD168 pKa = 3.15LCATFSRR175 pKa = 11.84NEE177 pKa = 4.28SINPITGKK185 pKa = 10.6KK186 pKa = 10.32LIGTSPIRR194 pKa = 11.84KK195 pKa = 8.42AWHH198 pKa = 6.02RR199 pKa = 11.84MCAGTPNTRR208 pKa = 11.84ATKK211 pKa = 10.57CMAFDD216 pKa = 4.94KK217 pKa = 10.78NDD219 pKa = 3.44KK220 pKa = 10.4KK221 pKa = 11.18NPFTGRR227 pKa = 11.84SIRR230 pKa = 11.84PEE232 pKa = 3.65QPAYY236 pKa = 10.69RR237 pKa = 11.84MVYY240 pKa = 9.71SMCHH244 pKa = 4.51GVPYY248 pKa = 10.35RR249 pKa = 11.84SPKK252 pKa = 7.71RR253 pKa = 11.84TRR255 pKa = 11.84RR256 pKa = 11.84SPVRR260 pKa = 11.84RR261 pKa = 11.84SPSPRR266 pKa = 11.84PYY268 pKa = 9.42TATSVTRR275 pKa = 11.84KK276 pKa = 8.58YY277 pKa = 10.96RR278 pKa = 11.84RR279 pKa = 11.84IKK281 pKa = 8.95TPARR285 pKa = 11.84SRR287 pKa = 11.84SRR289 pKa = 11.84SRR291 pKa = 11.84SNSVGRR297 pKa = 11.84RR298 pKa = 11.84RR299 pKa = 11.84TTAVKK304 pKa = 10.56SRR306 pKa = 11.84TKK308 pKa = 10.71SPARR312 pKa = 11.84RR313 pKa = 11.84QSVARR318 pKa = 11.84SRR320 pKa = 11.84SRR322 pKa = 11.84SKK324 pKa = 10.99SPVRR328 pKa = 11.84KK329 pKa = 5.63TTRR332 pKa = 11.84SRR334 pKa = 11.84TKK336 pKa = 10.25SPARR340 pKa = 11.84RR341 pKa = 11.84QSVARR346 pKa = 11.84SRR348 pKa = 11.84SRR350 pKa = 11.84TKK352 pKa = 10.66SPARR356 pKa = 11.84RR357 pKa = 11.84QSVARR362 pKa = 11.84SRR364 pKa = 11.84SRR366 pKa = 11.84SKK368 pKa = 10.93SPARR372 pKa = 11.84RR373 pKa = 11.84QSVARR378 pKa = 11.84SRR380 pKa = 11.84SRR382 pKa = 11.84SKK384 pKa = 10.93SPARR388 pKa = 11.84RR389 pKa = 11.84QSVAKK394 pKa = 10.05SRR396 pKa = 11.84SRR398 pKa = 11.84SKK400 pKa = 10.95SPARR404 pKa = 11.84RR405 pKa = 11.84QSVAMSRR412 pKa = 11.84SRR414 pKa = 11.84SRR416 pKa = 11.84SRR418 pKa = 11.84SRR420 pKa = 11.84QPMTAMRR427 pKa = 11.84RR428 pKa = 11.84STSRR432 pKa = 11.84ARR434 pKa = 11.84SRR436 pKa = 11.84SKK438 pKa = 10.59SRR440 pKa = 11.84SRR442 pKa = 11.84KK443 pKa = 9.7AMTASRR449 pKa = 11.84SRR451 pKa = 11.84SRR453 pKa = 11.84SVSRR457 pKa = 11.84YY458 pKa = 7.72MINPNFVPTTTKK470 pKa = 10.14KK471 pKa = 10.55KK472 pKa = 9.82RR473 pKa = 11.84VSPVSRR479 pKa = 11.84GRR481 pKa = 11.84INNSRR486 pKa = 11.84SRR488 pKa = 11.84SASRR492 pKa = 11.84ARR494 pKa = 11.84SGGLSPYY501 pKa = 9.82RR502 pKa = 11.84GRR504 pKa = 11.84VLLSPIPDD512 pKa = 3.73GATPMSRR519 pKa = 11.84SQLINIANNMNIAEE533 pKa = 4.24LRR535 pKa = 11.84HH536 pKa = 5.35IVVSNGFQPVRR547 pKa = 11.84VAQNTTQSQLLNIVKK562 pKa = 9.21FQIRR566 pKa = 11.84EE567 pKa = 4.25GNLKK571 pKa = 9.24WLPRR575 pKa = 11.84NDD577 pKa = 3.38QDD579 pKa = 3.62VPTYY583 pKa = 8.27YY584 pKa = 10.27TSSRR588 pKa = 11.84PFADD592 pKa = 3.04RR593 pKa = 11.84MKK595 pKa = 11.04KK596 pKa = 9.43NN597 pKa = 3.25
Molecular weight: 67.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
53577 |
53 |
1387 |
301.0 |
34.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.381 ± 0.162 | 2.684 ± 0.125 |
6.594 ± 0.126 | 5.168 ± 0.154 |
3.438 ± 0.116 | 4.789 ± 0.16 |
2.486 ± 0.086 | 5.417 ± 0.112 |
5.222 ± 0.155 | 8.27 ± 0.165 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.83 ± 0.06 | 5.411 ± 0.124 |
3.733 ± 0.123 | 2.815 ± 0.12 |
7.294 ± 0.227 | 8.031 ± 0.188 |
6.581 ± 0.136 | 7.813 ± 0.135 |
0.98 ± 0.067 | 4.061 ± 0.126 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
