
Pelotomaculum schinkii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Peptococcaceae; Pelotomaculum
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
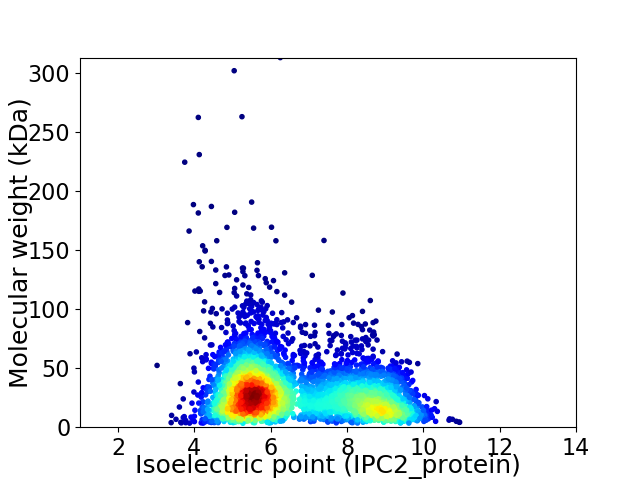
Virtual 2D-PAGE plot for 4132 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y7RCE5|A0A4Y7RCE5_9FIRM Stage III sporulation protein AE OS=Pelotomaculum schinkii OX=78350 GN=spoIIIAE PE=4 SV=1
MM1 pKa = 7.67RR2 pKa = 11.84SVLRR6 pKa = 11.84KK7 pKa = 8.94ILSMLLVLCLWTALMPFSVLAAEE30 pKa = 4.78GDD32 pKa = 3.44ICEE35 pKa = 4.56VNGTTYY41 pKa = 10.29ATLDD45 pKa = 3.64DD46 pKa = 5.08ALDD49 pKa = 4.05AVHH52 pKa = 6.54PTGGGTIRR60 pKa = 11.84LLEE63 pKa = 4.54NINYY67 pKa = 10.2DD68 pKa = 3.18GGIVIDD74 pKa = 4.04RR75 pKa = 11.84EE76 pKa = 4.65GITFDD81 pKa = 5.14LDD83 pKa = 3.72GFTLNVDD90 pKa = 3.24NSTGPGLKK98 pKa = 10.28VIGGGTVEE106 pKa = 5.83FIDD109 pKa = 4.16EE110 pKa = 4.29GTTGSFNVTGTEE122 pKa = 3.9YY123 pKa = 11.04GVWADD128 pKa = 3.56YY129 pKa = 11.38GSVVWVNNVTASGEE143 pKa = 4.27SSCGVKK149 pKa = 9.26ATEE152 pKa = 3.89YY153 pKa = 11.09ASIIVEE159 pKa = 4.18GNVIATNTNSIGVGVEE175 pKa = 3.73SGGEE179 pKa = 4.1VNVKK183 pKa = 10.66GDD185 pKa = 3.32VTANGIEE192 pKa = 4.53GIGIKK197 pKa = 10.22AYY199 pKa = 10.56DD200 pKa = 3.9VVCDD204 pKa = 3.52EE205 pKa = 5.2AEE207 pKa = 4.29VIVEE211 pKa = 4.14GDD213 pKa = 3.5VTASGGDD220 pKa = 3.62SVGVLAIDD228 pKa = 3.68SYY230 pKa = 10.36VTVGGNVTGDD240 pKa = 3.16KK241 pKa = 10.96GGVDD245 pKa = 3.89AEE247 pKa = 4.49DD248 pKa = 3.43SGIEE252 pKa = 3.76ITGDD256 pKa = 3.43VQSSDD261 pKa = 2.91EE262 pKa = 4.11SGYY265 pKa = 10.77GIRR268 pKa = 11.84ATDD271 pKa = 3.35NCDD274 pKa = 2.6IDD276 pKa = 3.98INGNVRR282 pKa = 11.84SNGTGVWIWAEE293 pKa = 4.05GTADD297 pKa = 3.7FSEE300 pKa = 4.09ITIDD304 pKa = 3.88GVIDD308 pKa = 3.69APEE311 pKa = 4.15YY312 pKa = 10.41IRR314 pKa = 11.84IGIDD318 pKa = 2.89TFDD321 pKa = 4.62FEE323 pKa = 6.57DD324 pKa = 5.53GYY326 pKa = 11.27LDD328 pKa = 4.22PDD330 pKa = 3.95MIDD333 pKa = 3.56GYY335 pKa = 10.59RR336 pKa = 11.84IYY338 pKa = 11.18VDD340 pKa = 3.56EE341 pKa = 4.47SLGAVLVAEE350 pKa = 4.81FAGGSGSSEE359 pKa = 3.66EE360 pKa = 4.42DD361 pKa = 3.62PYY363 pKa = 11.69LVAHH367 pKa = 7.34AAQLYY372 pKa = 8.69NVRR375 pKa = 11.84NHH377 pKa = 6.57LDD379 pKa = 3.06KK380 pKa = 10.97HH381 pKa = 6.02FKK383 pKa = 10.49LIEE386 pKa = 5.06DD387 pKa = 4.15IDD389 pKa = 4.23LSGYY393 pKa = 8.05STGDD397 pKa = 2.63GWEE400 pKa = 4.56PIGDD404 pKa = 4.15NIDD407 pKa = 3.82PFTGTFDD414 pKa = 3.6GGEE417 pKa = 3.93EE418 pKa = 5.33GYY420 pKa = 9.12TISGLVIDD428 pKa = 6.15RR429 pKa = 11.84ITTAYY434 pKa = 10.31VGLFGCTGAEE444 pKa = 3.83AEE446 pKa = 4.48IRR448 pKa = 11.84DD449 pKa = 4.44LRR451 pKa = 11.84LADD454 pKa = 3.7VNVTGSVYY462 pKa = 10.82VGGLVGRR469 pKa = 11.84NYY471 pKa = 10.89GQVTNSYY478 pKa = 8.96ATGDD482 pKa = 3.33VTARR486 pKa = 11.84SDD488 pKa = 3.29GGGLVGQNEE497 pKa = 4.3GSITDD502 pKa = 3.79SYY504 pKa = 11.81AIGTVTDD511 pKa = 3.31VWSGTFGGLVGHH523 pKa = 6.68NNGTITNCHH532 pKa = 6.3AEE534 pKa = 4.27VTVKK538 pKa = 10.79SNEE541 pKa = 4.02SMVGGLVGEE550 pKa = 4.16NDD552 pKa = 3.65GGISEE557 pKa = 4.77SYY559 pKa = 10.94AVGEE563 pKa = 4.28VEE565 pKa = 4.61GKK567 pKa = 10.19SEE569 pKa = 3.98VGGLVGYY576 pKa = 10.69NSGGNIDD583 pKa = 3.76RR584 pKa = 11.84SYY586 pKa = 11.03ATGAVTGTEE595 pKa = 3.59TGIYY599 pKa = 7.24YY600 pKa = 9.34TYY602 pKa = 11.09VGGLVGINAGPISEE616 pKa = 4.4SYY618 pKa = 11.17ARR620 pKa = 11.84GAVSGQEE627 pKa = 4.19DD628 pKa = 3.25IGGLVGYY635 pKa = 10.03NSGSGSVTNSYY646 pKa = 9.1ATGTVNGYY654 pKa = 10.4SYY656 pKa = 10.93IGGLAGLNLGNIMDD670 pKa = 4.79SYY672 pKa = 11.01WDD674 pKa = 3.87TEE676 pKa = 4.15TSLQRR681 pKa = 11.84SSDD684 pKa = 3.32GGTGKK689 pKa = 8.75ATAEE693 pKa = 4.26MKK695 pKa = 10.13QQATFTDD702 pKa = 3.34WDD704 pKa = 4.26FDD706 pKa = 4.76TIWEE710 pKa = 4.51INGSDD715 pKa = 3.44NNGYY719 pKa = 9.31PFLRR723 pKa = 11.84WQGYY727 pKa = 10.21DD728 pKa = 2.9GGQTNEE734 pKa = 4.27PPVLSAVGVSAVTQTTATLNFTSDD758 pKa = 2.94KK759 pKa = 10.88AGNYY763 pKa = 9.17YY764 pKa = 10.48FLVYY768 pKa = 10.41SAGDD772 pKa = 3.89DD773 pKa = 3.81APDD776 pKa = 3.59AATIKK781 pKa = 10.77AQGEE785 pKa = 4.0AAAKK789 pKa = 8.53GTGTAAAAANAAQVTGLTASTAYY812 pKa = 8.98KK813 pKa = 10.28AYY815 pKa = 10.8VIVEE819 pKa = 4.15DD820 pKa = 4.11AEE822 pKa = 4.47EE823 pKa = 4.48NISNVAVIGFTTATAEE839 pKa = 4.12MPIGADD845 pKa = 3.34ISPALCTFDD854 pKa = 5.86LEE856 pKa = 4.85NPDD859 pKa = 3.76NVSTTISWGSATTVTAVVYY878 pKa = 7.7GTTSLTTPADD888 pKa = 3.75YY889 pKa = 10.83NVSGNTLIITAGYY902 pKa = 10.52LEE904 pKa = 4.58GLNLSAGDD912 pKa = 3.65TVGFEE917 pKa = 3.86ISFDD921 pKa = 3.84VGDD924 pKa = 4.16SASLTVNIVNGYY936 pKa = 9.7IPGTDD941 pKa = 3.12ATLSDD946 pKa = 4.14LKK948 pKa = 11.36VGGVTVAGFVYY959 pKa = 10.62DD960 pKa = 3.61NYY962 pKa = 10.98DD963 pKa = 3.19YY964 pKa = 11.07SIQLPYY970 pKa = 9.55GTLPGSLAATVSATPNDD987 pKa = 3.95PKK989 pKa = 11.06AGVAITQAASLPGSATIVVTAEE1011 pKa = 3.9DD1012 pKa = 3.72TMTTQTYY1019 pKa = 7.92TVNFTLEE1026 pKa = 4.07AASNNAPNRR1035 pKa = 11.84KK1036 pKa = 7.47TGVPATTSASVTVNTAYY1053 pKa = 9.69TLDD1056 pKa = 3.75LSTIFEE1062 pKa = 4.91DD1063 pKa = 4.8ADD1065 pKa = 4.12SDD1067 pKa = 4.13PLTYY1071 pKa = 9.84TVSIDD1076 pKa = 3.39GANPVAAAANYY1087 pKa = 10.2VYY1089 pKa = 10.13TPTSAGDD1096 pKa = 3.49TILVFKK1102 pKa = 11.02ANDD1105 pKa = 3.64GTADD1109 pKa = 3.45STDD1112 pKa = 3.55TYY1114 pKa = 11.2TVTLTANAAPPQTYY1128 pKa = 10.75ALTITAGTGGSITTGSSGNYY1148 pKa = 9.17AAGTVISIAAAPSSGYY1164 pKa = 10.71SFSKK1168 pKa = 9.32WSSVGGGTFGSTTSASTTFTMPAGAVTITAGFTHH1202 pKa = 6.57NGSGGGNSGGGGSTTPSTPTYY1223 pKa = 9.96KK1224 pKa = 10.7ADD1226 pKa = 3.3VKK1228 pKa = 10.88AGNGSEE1234 pKa = 4.12TTLPVTVDD1242 pKa = 3.25KK1243 pKa = 11.32DD1244 pKa = 3.29AGTASIDD1251 pKa = 3.71ADD1253 pKa = 4.01SQSLAQGGTVITIPSIPDD1271 pKa = 2.92IDD1273 pKa = 4.2TYY1275 pKa = 11.69SLGIPVPDD1283 pKa = 4.84LSTTDD1288 pKa = 3.38VQGTLSFNTDD1298 pKa = 2.89AVSITVPSNMLTGVADD1314 pKa = 3.86ADD1316 pKa = 3.93GNRR1319 pKa = 11.84AQITIGQGDD1328 pKa = 4.15KK1329 pKa = 11.41SNLSEE1334 pKa = 4.22DD1335 pKa = 3.66VKK1337 pKa = 11.19DD1338 pKa = 4.0AVGDD1342 pKa = 3.78RR1343 pKa = 11.84PLVQLTLSIDD1353 pKa = 3.86GKK1355 pKa = 7.37QTNWNNPDD1363 pKa = 3.27APVTVSIPYY1372 pKa = 9.03TPTAAEE1378 pKa = 3.86LAAPEE1383 pKa = 5.09HH1384 pKa = 5.95IVVWYY1389 pKa = 10.22INGAGNAVSVPNGRR1403 pKa = 11.84YY1404 pKa = 9.77DD1405 pKa = 3.63PATGAVTFTTTHH1417 pKa = 5.99FSYY1420 pKa = 10.95YY1421 pKa = 10.46AIAYY1425 pKa = 6.1VHH1427 pKa = 6.13KK1428 pKa = 9.39TFDD1431 pKa = 4.05DD1432 pKa = 4.23LEE1434 pKa = 4.4SVAWAKK1440 pKa = 10.74EE1441 pKa = 4.21SIEE1444 pKa = 3.97VLASKK1449 pKa = 10.82GILEE1453 pKa = 5.28GISEE1457 pKa = 4.58TEE1459 pKa = 3.81YY1460 pKa = 11.18SPQEE1464 pKa = 4.29NITRR1468 pKa = 11.84ADD1470 pKa = 3.4FLFFLVRR1477 pKa = 11.84TLGVDD1482 pKa = 3.33AEE1484 pKa = 4.74VNGNFDD1490 pKa = 5.65DD1491 pKa = 4.21ISSDD1495 pKa = 2.85AYY1497 pKa = 9.37YY1498 pKa = 10.94NKK1500 pKa = 10.3EE1501 pKa = 3.35IVIAKK1506 pKa = 9.95KK1507 pKa = 10.65LGITGGTGDD1516 pKa = 4.32NKK1518 pKa = 10.21FSPDD1522 pKa = 3.21ACITRR1527 pKa = 11.84QDD1529 pKa = 3.51MMVLTEE1535 pKa = 3.82RR1536 pKa = 11.84ALRR1539 pKa = 11.84MLKK1542 pKa = 10.33KK1543 pKa = 10.68LEE1545 pKa = 4.27AQGSASDD1552 pKa = 3.92LVRR1555 pKa = 11.84FTDD1558 pKa = 3.32KK1559 pKa = 11.29SLVAAYY1565 pKa = 9.54AVNSVASVIKK1575 pKa = 10.12EE1576 pKa = 3.71GLIVGSGDD1584 pKa = 3.98KK1585 pKa = 10.56INPLGNTTRR1594 pKa = 11.84AEE1596 pKa = 4.05ATVFLYY1602 pKa = 9.89RR1603 pKa = 11.84VYY1605 pKa = 11.31NKK1607 pKa = 9.15YY1608 pKa = 9.11TGYY1611 pKa = 11.3
MM1 pKa = 7.67RR2 pKa = 11.84SVLRR6 pKa = 11.84KK7 pKa = 8.94ILSMLLVLCLWTALMPFSVLAAEE30 pKa = 4.78GDD32 pKa = 3.44ICEE35 pKa = 4.56VNGTTYY41 pKa = 10.29ATLDD45 pKa = 3.64DD46 pKa = 5.08ALDD49 pKa = 4.05AVHH52 pKa = 6.54PTGGGTIRR60 pKa = 11.84LLEE63 pKa = 4.54NINYY67 pKa = 10.2DD68 pKa = 3.18GGIVIDD74 pKa = 4.04RR75 pKa = 11.84EE76 pKa = 4.65GITFDD81 pKa = 5.14LDD83 pKa = 3.72GFTLNVDD90 pKa = 3.24NSTGPGLKK98 pKa = 10.28VIGGGTVEE106 pKa = 5.83FIDD109 pKa = 4.16EE110 pKa = 4.29GTTGSFNVTGTEE122 pKa = 3.9YY123 pKa = 11.04GVWADD128 pKa = 3.56YY129 pKa = 11.38GSVVWVNNVTASGEE143 pKa = 4.27SSCGVKK149 pKa = 9.26ATEE152 pKa = 3.89YY153 pKa = 11.09ASIIVEE159 pKa = 4.18GNVIATNTNSIGVGVEE175 pKa = 3.73SGGEE179 pKa = 4.1VNVKK183 pKa = 10.66GDD185 pKa = 3.32VTANGIEE192 pKa = 4.53GIGIKK197 pKa = 10.22AYY199 pKa = 10.56DD200 pKa = 3.9VVCDD204 pKa = 3.52EE205 pKa = 5.2AEE207 pKa = 4.29VIVEE211 pKa = 4.14GDD213 pKa = 3.5VTASGGDD220 pKa = 3.62SVGVLAIDD228 pKa = 3.68SYY230 pKa = 10.36VTVGGNVTGDD240 pKa = 3.16KK241 pKa = 10.96GGVDD245 pKa = 3.89AEE247 pKa = 4.49DD248 pKa = 3.43SGIEE252 pKa = 3.76ITGDD256 pKa = 3.43VQSSDD261 pKa = 2.91EE262 pKa = 4.11SGYY265 pKa = 10.77GIRR268 pKa = 11.84ATDD271 pKa = 3.35NCDD274 pKa = 2.6IDD276 pKa = 3.98INGNVRR282 pKa = 11.84SNGTGVWIWAEE293 pKa = 4.05GTADD297 pKa = 3.7FSEE300 pKa = 4.09ITIDD304 pKa = 3.88GVIDD308 pKa = 3.69APEE311 pKa = 4.15YY312 pKa = 10.41IRR314 pKa = 11.84IGIDD318 pKa = 2.89TFDD321 pKa = 4.62FEE323 pKa = 6.57DD324 pKa = 5.53GYY326 pKa = 11.27LDD328 pKa = 4.22PDD330 pKa = 3.95MIDD333 pKa = 3.56GYY335 pKa = 10.59RR336 pKa = 11.84IYY338 pKa = 11.18VDD340 pKa = 3.56EE341 pKa = 4.47SLGAVLVAEE350 pKa = 4.81FAGGSGSSEE359 pKa = 3.66EE360 pKa = 4.42DD361 pKa = 3.62PYY363 pKa = 11.69LVAHH367 pKa = 7.34AAQLYY372 pKa = 8.69NVRR375 pKa = 11.84NHH377 pKa = 6.57LDD379 pKa = 3.06KK380 pKa = 10.97HH381 pKa = 6.02FKK383 pKa = 10.49LIEE386 pKa = 5.06DD387 pKa = 4.15IDD389 pKa = 4.23LSGYY393 pKa = 8.05STGDD397 pKa = 2.63GWEE400 pKa = 4.56PIGDD404 pKa = 4.15NIDD407 pKa = 3.82PFTGTFDD414 pKa = 3.6GGEE417 pKa = 3.93EE418 pKa = 5.33GYY420 pKa = 9.12TISGLVIDD428 pKa = 6.15RR429 pKa = 11.84ITTAYY434 pKa = 10.31VGLFGCTGAEE444 pKa = 3.83AEE446 pKa = 4.48IRR448 pKa = 11.84DD449 pKa = 4.44LRR451 pKa = 11.84LADD454 pKa = 3.7VNVTGSVYY462 pKa = 10.82VGGLVGRR469 pKa = 11.84NYY471 pKa = 10.89GQVTNSYY478 pKa = 8.96ATGDD482 pKa = 3.33VTARR486 pKa = 11.84SDD488 pKa = 3.29GGGLVGQNEE497 pKa = 4.3GSITDD502 pKa = 3.79SYY504 pKa = 11.81AIGTVTDD511 pKa = 3.31VWSGTFGGLVGHH523 pKa = 6.68NNGTITNCHH532 pKa = 6.3AEE534 pKa = 4.27VTVKK538 pKa = 10.79SNEE541 pKa = 4.02SMVGGLVGEE550 pKa = 4.16NDD552 pKa = 3.65GGISEE557 pKa = 4.77SYY559 pKa = 10.94AVGEE563 pKa = 4.28VEE565 pKa = 4.61GKK567 pKa = 10.19SEE569 pKa = 3.98VGGLVGYY576 pKa = 10.69NSGGNIDD583 pKa = 3.76RR584 pKa = 11.84SYY586 pKa = 11.03ATGAVTGTEE595 pKa = 3.59TGIYY599 pKa = 7.24YY600 pKa = 9.34TYY602 pKa = 11.09VGGLVGINAGPISEE616 pKa = 4.4SYY618 pKa = 11.17ARR620 pKa = 11.84GAVSGQEE627 pKa = 4.19DD628 pKa = 3.25IGGLVGYY635 pKa = 10.03NSGSGSVTNSYY646 pKa = 9.1ATGTVNGYY654 pKa = 10.4SYY656 pKa = 10.93IGGLAGLNLGNIMDD670 pKa = 4.79SYY672 pKa = 11.01WDD674 pKa = 3.87TEE676 pKa = 4.15TSLQRR681 pKa = 11.84SSDD684 pKa = 3.32GGTGKK689 pKa = 8.75ATAEE693 pKa = 4.26MKK695 pKa = 10.13QQATFTDD702 pKa = 3.34WDD704 pKa = 4.26FDD706 pKa = 4.76TIWEE710 pKa = 4.51INGSDD715 pKa = 3.44NNGYY719 pKa = 9.31PFLRR723 pKa = 11.84WQGYY727 pKa = 10.21DD728 pKa = 2.9GGQTNEE734 pKa = 4.27PPVLSAVGVSAVTQTTATLNFTSDD758 pKa = 2.94KK759 pKa = 10.88AGNYY763 pKa = 9.17YY764 pKa = 10.48FLVYY768 pKa = 10.41SAGDD772 pKa = 3.89DD773 pKa = 3.81APDD776 pKa = 3.59AATIKK781 pKa = 10.77AQGEE785 pKa = 4.0AAAKK789 pKa = 8.53GTGTAAAAANAAQVTGLTASTAYY812 pKa = 8.98KK813 pKa = 10.28AYY815 pKa = 10.8VIVEE819 pKa = 4.15DD820 pKa = 4.11AEE822 pKa = 4.47EE823 pKa = 4.48NISNVAVIGFTTATAEE839 pKa = 4.12MPIGADD845 pKa = 3.34ISPALCTFDD854 pKa = 5.86LEE856 pKa = 4.85NPDD859 pKa = 3.76NVSTTISWGSATTVTAVVYY878 pKa = 7.7GTTSLTTPADD888 pKa = 3.75YY889 pKa = 10.83NVSGNTLIITAGYY902 pKa = 10.52LEE904 pKa = 4.58GLNLSAGDD912 pKa = 3.65TVGFEE917 pKa = 3.86ISFDD921 pKa = 3.84VGDD924 pKa = 4.16SASLTVNIVNGYY936 pKa = 9.7IPGTDD941 pKa = 3.12ATLSDD946 pKa = 4.14LKK948 pKa = 11.36VGGVTVAGFVYY959 pKa = 10.62DD960 pKa = 3.61NYY962 pKa = 10.98DD963 pKa = 3.19YY964 pKa = 11.07SIQLPYY970 pKa = 9.55GTLPGSLAATVSATPNDD987 pKa = 3.95PKK989 pKa = 11.06AGVAITQAASLPGSATIVVTAEE1011 pKa = 3.9DD1012 pKa = 3.72TMTTQTYY1019 pKa = 7.92TVNFTLEE1026 pKa = 4.07AASNNAPNRR1035 pKa = 11.84KK1036 pKa = 7.47TGVPATTSASVTVNTAYY1053 pKa = 9.69TLDD1056 pKa = 3.75LSTIFEE1062 pKa = 4.91DD1063 pKa = 4.8ADD1065 pKa = 4.12SDD1067 pKa = 4.13PLTYY1071 pKa = 9.84TVSIDD1076 pKa = 3.39GANPVAAAANYY1087 pKa = 10.2VYY1089 pKa = 10.13TPTSAGDD1096 pKa = 3.49TILVFKK1102 pKa = 11.02ANDD1105 pKa = 3.64GTADD1109 pKa = 3.45STDD1112 pKa = 3.55TYY1114 pKa = 11.2TVTLTANAAPPQTYY1128 pKa = 10.75ALTITAGTGGSITTGSSGNYY1148 pKa = 9.17AAGTVISIAAAPSSGYY1164 pKa = 10.71SFSKK1168 pKa = 9.32WSSVGGGTFGSTTSASTTFTMPAGAVTITAGFTHH1202 pKa = 6.57NGSGGGNSGGGGSTTPSTPTYY1223 pKa = 9.96KK1224 pKa = 10.7ADD1226 pKa = 3.3VKK1228 pKa = 10.88AGNGSEE1234 pKa = 4.12TTLPVTVDD1242 pKa = 3.25KK1243 pKa = 11.32DD1244 pKa = 3.29AGTASIDD1251 pKa = 3.71ADD1253 pKa = 4.01SQSLAQGGTVITIPSIPDD1271 pKa = 2.92IDD1273 pKa = 4.2TYY1275 pKa = 11.69SLGIPVPDD1283 pKa = 4.84LSTTDD1288 pKa = 3.38VQGTLSFNTDD1298 pKa = 2.89AVSITVPSNMLTGVADD1314 pKa = 3.86ADD1316 pKa = 3.93GNRR1319 pKa = 11.84AQITIGQGDD1328 pKa = 4.15KK1329 pKa = 11.41SNLSEE1334 pKa = 4.22DD1335 pKa = 3.66VKK1337 pKa = 11.19DD1338 pKa = 4.0AVGDD1342 pKa = 3.78RR1343 pKa = 11.84PLVQLTLSIDD1353 pKa = 3.86GKK1355 pKa = 7.37QTNWNNPDD1363 pKa = 3.27APVTVSIPYY1372 pKa = 9.03TPTAAEE1378 pKa = 3.86LAAPEE1383 pKa = 5.09HH1384 pKa = 5.95IVVWYY1389 pKa = 10.22INGAGNAVSVPNGRR1403 pKa = 11.84YY1404 pKa = 9.77DD1405 pKa = 3.63PATGAVTFTTTHH1417 pKa = 5.99FSYY1420 pKa = 10.95YY1421 pKa = 10.46AIAYY1425 pKa = 6.1VHH1427 pKa = 6.13KK1428 pKa = 9.39TFDD1431 pKa = 4.05DD1432 pKa = 4.23LEE1434 pKa = 4.4SVAWAKK1440 pKa = 10.74EE1441 pKa = 4.21SIEE1444 pKa = 3.97VLASKK1449 pKa = 10.82GILEE1453 pKa = 5.28GISEE1457 pKa = 4.58TEE1459 pKa = 3.81YY1460 pKa = 11.18SPQEE1464 pKa = 4.29NITRR1468 pKa = 11.84ADD1470 pKa = 3.4FLFFLVRR1477 pKa = 11.84TLGVDD1482 pKa = 3.33AEE1484 pKa = 4.74VNGNFDD1490 pKa = 5.65DD1491 pKa = 4.21ISSDD1495 pKa = 2.85AYY1497 pKa = 9.37YY1498 pKa = 10.94NKK1500 pKa = 10.3EE1501 pKa = 3.35IVIAKK1506 pKa = 9.95KK1507 pKa = 10.65LGITGGTGDD1516 pKa = 4.32NKK1518 pKa = 10.21FSPDD1522 pKa = 3.21ACITRR1527 pKa = 11.84QDD1529 pKa = 3.51MMVLTEE1535 pKa = 3.82RR1536 pKa = 11.84ALRR1539 pKa = 11.84MLKK1542 pKa = 10.33KK1543 pKa = 10.68LEE1545 pKa = 4.27AQGSASDD1552 pKa = 3.92LVRR1555 pKa = 11.84FTDD1558 pKa = 3.32KK1559 pKa = 11.29SLVAAYY1565 pKa = 9.54AVNSVASVIKK1575 pKa = 10.12EE1576 pKa = 3.71GLIVGSGDD1584 pKa = 3.98KK1585 pKa = 10.56INPLGNTTRR1594 pKa = 11.84AEE1596 pKa = 4.05ATVFLYY1602 pKa = 9.89RR1603 pKa = 11.84VYY1605 pKa = 11.31NKK1607 pKa = 9.15YY1608 pKa = 9.11TGYY1611 pKa = 11.3
Molecular weight: 166.1 kDa
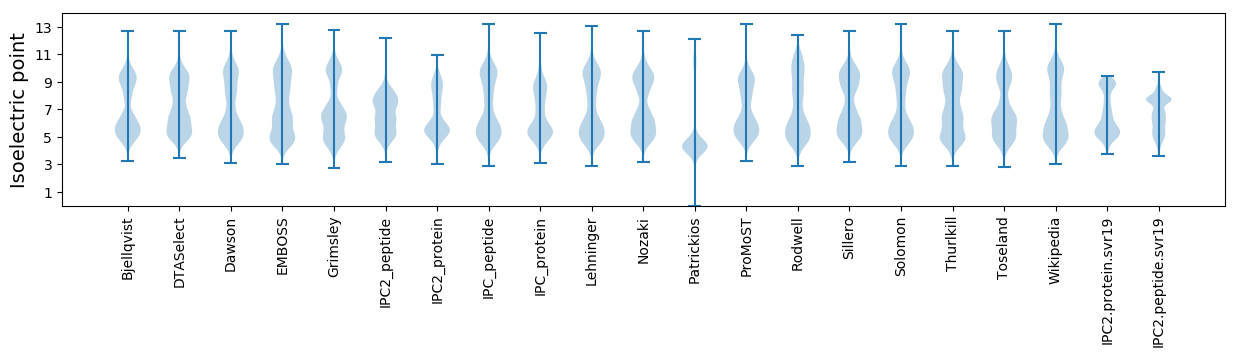
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y7RD59|A0A4Y7RD59_9FIRM Formate hydrogenlyase subunit 7 OS=Pelotomaculum schinkii OX=78350 GN=hycG_1 PE=4 SV=1
MM1 pKa = 6.95KK2 pKa = 10.11QRR4 pKa = 11.84RR5 pKa = 11.84TSLRR9 pKa = 11.84PMRR12 pKa = 11.84MKK14 pKa = 10.39RR15 pKa = 11.84VLIIAGIMATIARR28 pKa = 11.84IQVNGLQGG36 pKa = 3.14
MM1 pKa = 6.95KK2 pKa = 10.11QRR4 pKa = 11.84RR5 pKa = 11.84TSLRR9 pKa = 11.84PMRR12 pKa = 11.84MKK14 pKa = 10.39RR15 pKa = 11.84VLIIAGIMATIARR28 pKa = 11.84IQVNGLQGG36 pKa = 3.14
Molecular weight: 4.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1213829 |
30 |
2778 |
293.8 |
32.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.635 ± 0.047 | 1.274 ± 0.021 |
5.027 ± 0.028 | 6.639 ± 0.044 |
3.868 ± 0.028 | 7.908 ± 0.047 |
1.725 ± 0.017 | 6.794 ± 0.035 |
5.794 ± 0.038 | 10.139 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.56 ± 0.018 | 4.025 ± 0.033 |
4.242 ± 0.029 | 3.393 ± 0.024 |
5.243 ± 0.039 | 5.699 ± 0.033 |
5.257 ± 0.044 | 7.423 ± 0.035 |
1.018 ± 0.015 | 3.339 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
