
Streptomyces showdoensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
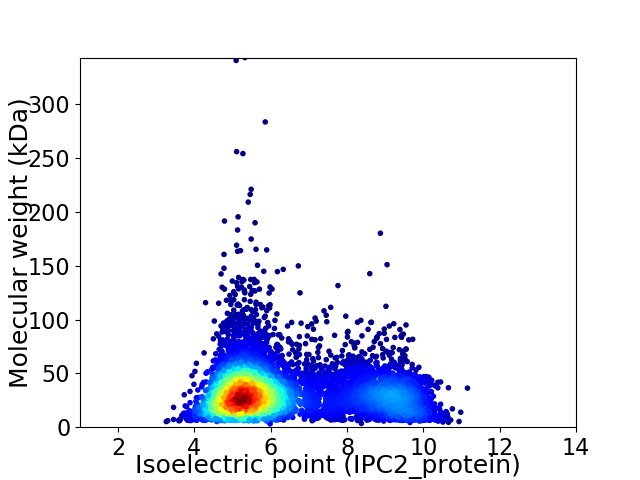
Virtual 2D-PAGE plot for 6933 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P2GW93|A0A2P2GW93_9ACTN Glutamate--tRNA ligase OS=Streptomyces showdoensis OX=68268 GN=gltX PE=3 SV=1
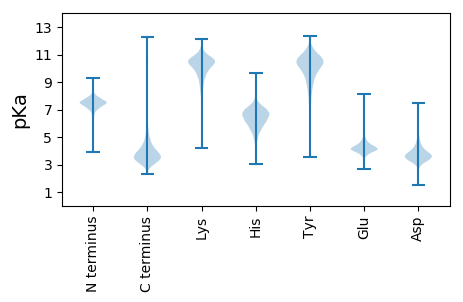
MM1 pKa = 7.15ITTDD5 pKa = 4.28FSPGSPCWLDD15 pKa = 3.92LGAPDD20 pKa = 4.22VPAAAAFYY28 pKa = 10.05GAVFGWEE35 pKa = 4.16YY36 pKa = 11.23EE37 pKa = 4.32SMDD40 pKa = 3.55GDD42 pKa = 3.82EE43 pKa = 4.96GGGGAEE49 pKa = 4.29GTDD52 pKa = 3.46AGSGADD58 pKa = 3.62SGAEE62 pKa = 3.89NTGTTEE68 pKa = 3.95DD69 pKa = 4.11FEE71 pKa = 6.91GGMFRR76 pKa = 11.84KK77 pKa = 10.06DD78 pKa = 3.17GRR80 pKa = 11.84IVAGLGSLTEE90 pKa = 4.07EE91 pKa = 4.92GARR94 pKa = 11.84SAWMIYY100 pKa = 10.45YY101 pKa = 10.44SVDD104 pKa = 3.33DD105 pKa = 4.6ADD107 pKa = 3.85ATTEE111 pKa = 3.92AVVQGGGTVRR121 pKa = 11.84VPPRR125 pKa = 11.84DD126 pKa = 3.29LDD128 pKa = 3.26EE129 pKa = 4.5WGRR132 pKa = 11.84MAQYY136 pKa = 10.81SDD138 pKa = 3.64PLGGQFAVWQPGSNKK153 pKa = 9.73GAEE156 pKa = 4.32LVDD159 pKa = 4.07EE160 pKa = 5.15PGSLSWTEE168 pKa = 5.02LYY170 pKa = 10.58TSDD173 pKa = 3.72AAAARR178 pKa = 11.84EE179 pKa = 4.55FYY181 pKa = 11.11ASVLGWWYY189 pKa = 11.39SDD191 pKa = 2.97MEE193 pKa = 4.37MPGGEE198 pKa = 4.25GTYY201 pKa = 10.11TLITPSGLPAEE212 pKa = 4.44RR213 pKa = 11.84MHH215 pKa = 7.16GGLLEE220 pKa = 4.69LSPDD224 pKa = 3.9DD225 pKa = 4.0LALAGGRR232 pKa = 11.84PYY234 pKa = 9.51WHH236 pKa = 7.23PVFAVTDD243 pKa = 4.04CDD245 pKa = 3.29ATAAAVTANGGSVQMGPADD264 pKa = 3.53VPGVGRR270 pKa = 11.84LAVCLDD276 pKa = 3.61PANADD281 pKa = 3.71FVVLAPEE288 pKa = 4.28ANGG291 pKa = 3.42
MM1 pKa = 7.15ITTDD5 pKa = 4.28FSPGSPCWLDD15 pKa = 3.92LGAPDD20 pKa = 4.22VPAAAAFYY28 pKa = 10.05GAVFGWEE35 pKa = 4.16YY36 pKa = 11.23EE37 pKa = 4.32SMDD40 pKa = 3.55GDD42 pKa = 3.82EE43 pKa = 4.96GGGGAEE49 pKa = 4.29GTDD52 pKa = 3.46AGSGADD58 pKa = 3.62SGAEE62 pKa = 3.89NTGTTEE68 pKa = 3.95DD69 pKa = 4.11FEE71 pKa = 6.91GGMFRR76 pKa = 11.84KK77 pKa = 10.06DD78 pKa = 3.17GRR80 pKa = 11.84IVAGLGSLTEE90 pKa = 4.07EE91 pKa = 4.92GARR94 pKa = 11.84SAWMIYY100 pKa = 10.45YY101 pKa = 10.44SVDD104 pKa = 3.33DD105 pKa = 4.6ADD107 pKa = 3.85ATTEE111 pKa = 3.92AVVQGGGTVRR121 pKa = 11.84VPPRR125 pKa = 11.84DD126 pKa = 3.29LDD128 pKa = 3.26EE129 pKa = 4.5WGRR132 pKa = 11.84MAQYY136 pKa = 10.81SDD138 pKa = 3.64PLGGQFAVWQPGSNKK153 pKa = 9.73GAEE156 pKa = 4.32LVDD159 pKa = 4.07EE160 pKa = 5.15PGSLSWTEE168 pKa = 5.02LYY170 pKa = 10.58TSDD173 pKa = 3.72AAAARR178 pKa = 11.84EE179 pKa = 4.55FYY181 pKa = 11.11ASVLGWWYY189 pKa = 11.39SDD191 pKa = 2.97MEE193 pKa = 4.37MPGGEE198 pKa = 4.25GTYY201 pKa = 10.11TLITPSGLPAEE212 pKa = 4.44RR213 pKa = 11.84MHH215 pKa = 7.16GGLLEE220 pKa = 4.69LSPDD224 pKa = 3.9DD225 pKa = 4.0LALAGGRR232 pKa = 11.84PYY234 pKa = 9.51WHH236 pKa = 7.23PVFAVTDD243 pKa = 4.04CDD245 pKa = 3.29ATAAAVTANGGSVQMGPADD264 pKa = 3.53VPGVGRR270 pKa = 11.84LAVCLDD276 pKa = 3.61PANADD281 pKa = 3.71FVVLAPEE288 pKa = 4.28ANGG291 pKa = 3.42
Molecular weight: 30.12 kDa
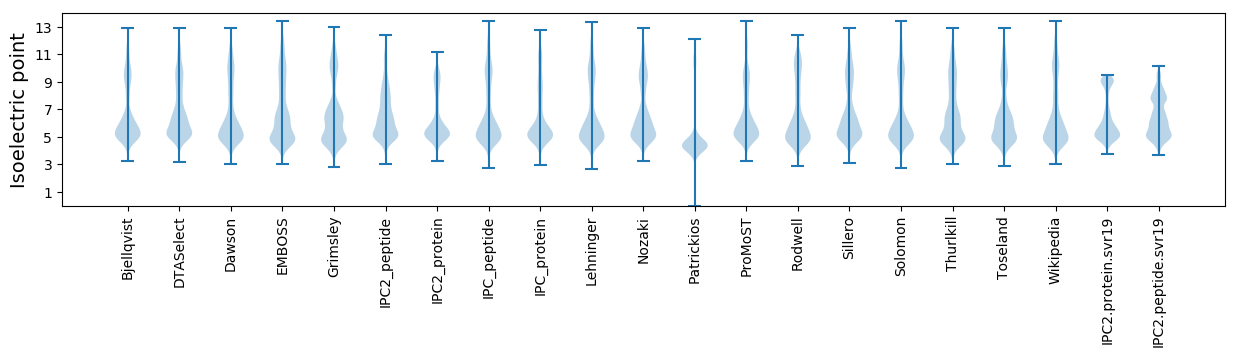
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P2GK66|A0A2P2GK66_9ACTN Alpha/beta hydrolase OS=Streptomyces showdoensis OX=68268 GN=VO63_20915 PE=4 SV=1
MM1 pKa = 6.5VAAVAVATVAVTSVRR16 pKa = 11.84RR17 pKa = 11.84PRR19 pKa = 11.84TSVTTAGTTTVAVAVATVAATTVPPSAVTTTAVVAAAAVSVGMTTVAAAAVAGSVATTGVTTTVVAAASAVTTVRR94 pKa = 11.84RR95 pKa = 11.84PRR97 pKa = 11.84TSVTTAGTTTAVAAVATVAATTVPPSVATTTAVVAAVAVATVAVTSVRR145 pKa = 11.84RR146 pKa = 11.84PRR148 pKa = 11.84TSVTTAGTTTVAVAVATVAATTVPPSAVTTTAVVAAVATVAVTSVRR194 pKa = 11.84RR195 pKa = 11.84PRR197 pKa = 11.84TSVTTAGTTTVVAAGSVATTGVTTTGAAAASAVTTAGTTTVVAASVATTVRR248 pKa = 11.84LSVATSAVTSVRR260 pKa = 11.84PPRR263 pKa = 11.84TSVTTAATSAPPRR276 pKa = 11.84TSVTTAATSAPPRR289 pKa = 11.84TSVTTAATTTAVAASVATTGVTSGPVVRR317 pKa = 11.84VVTTTGVVAAVSVAATTVGTTGAVSAAGTTVAGTGAGTTGTVRR360 pKa = 11.84TARR363 pKa = 11.84RR364 pKa = 11.84SSGCRR369 pKa = 11.84SRR371 pKa = 11.84RR372 pKa = 11.84RR373 pKa = 11.84SPVRR377 pKa = 11.84RR378 pKa = 11.84STRR381 pKa = 11.84MCARR385 pKa = 11.84SS386 pKa = 3.1
MM1 pKa = 6.5VAAVAVATVAVTSVRR16 pKa = 11.84RR17 pKa = 11.84PRR19 pKa = 11.84TSVTTAGTTTVAVAVATVAATTVPPSAVTTTAVVAAAAVSVGMTTVAAAAVAGSVATTGVTTTVVAAASAVTTVRR94 pKa = 11.84RR95 pKa = 11.84PRR97 pKa = 11.84TSVTTAGTTTAVAAVATVAATTVPPSVATTTAVVAAVAVATVAVTSVRR145 pKa = 11.84RR146 pKa = 11.84PRR148 pKa = 11.84TSVTTAGTTTVAVAVATVAATTVPPSAVTTTAVVAAVATVAVTSVRR194 pKa = 11.84RR195 pKa = 11.84PRR197 pKa = 11.84TSVTTAGTTTVVAAGSVATTGVTTTGAAAASAVTTAGTTTVVAASVATTVRR248 pKa = 11.84LSVATSAVTSVRR260 pKa = 11.84PPRR263 pKa = 11.84TSVTTAATSAPPRR276 pKa = 11.84TSVTTAATSAPPRR289 pKa = 11.84TSVTTAATTTAVAASVATTGVTSGPVVRR317 pKa = 11.84VVTTTGVVAAVSVAATTVGTTGAVSAAGTTVAGTGAGTTGTVRR360 pKa = 11.84TARR363 pKa = 11.84RR364 pKa = 11.84SSGCRR369 pKa = 11.84SRR371 pKa = 11.84RR372 pKa = 11.84RR373 pKa = 11.84SPVRR377 pKa = 11.84RR378 pKa = 11.84STRR381 pKa = 11.84MCARR385 pKa = 11.84SS386 pKa = 3.1
Molecular weight: 36.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2226963 |
29 |
3201 |
321.2 |
34.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.1 ± 0.047 | 0.761 ± 0.009 |
5.823 ± 0.024 | 5.795 ± 0.035 |
2.738 ± 0.017 | 9.733 ± 0.029 |
2.198 ± 0.012 | 2.991 ± 0.02 |
2.145 ± 0.023 | 10.462 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.679 ± 0.011 | 1.703 ± 0.016 |
6.219 ± 0.026 | 2.564 ± 0.02 |
8.035 ± 0.032 | 4.798 ± 0.02 |
6.132 ± 0.029 | 8.487 ± 0.03 |
1.527 ± 0.012 | 2.109 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
