
Sonchus yellow net virus (SYNV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
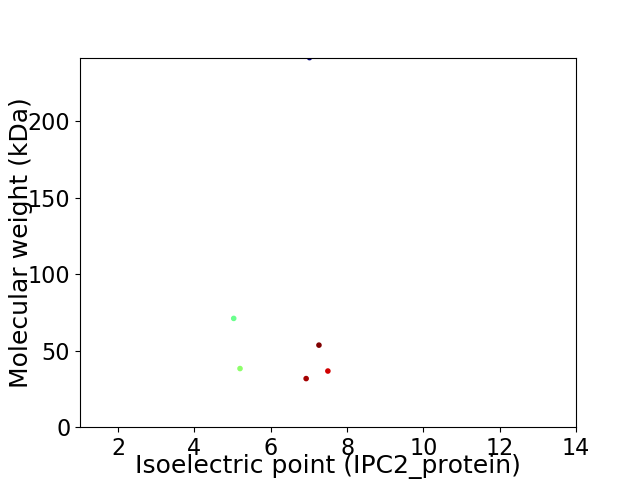
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|P31332|L_SYNV RNA-directed RNA polymerase L OS=Sonchus yellow net virus OX=11307 GN=L PE=2 SV=1
MM1 pKa = 7.07SHH3 pKa = 6.53IMNLLVISFVLAGSSWSLLGYY24 pKa = 10.25QDD26 pKa = 4.9DD27 pKa = 4.41FSSKK31 pKa = 10.19RR32 pKa = 11.84SGALASNPTYY42 pKa = 10.73NLPQDD47 pKa = 3.27KK48 pKa = 10.87GYY50 pKa = 10.71GRR52 pKa = 11.84DD53 pKa = 3.31MYY55 pKa = 10.5QPYY58 pKa = 9.85YY59 pKa = 10.01ICEE62 pKa = 4.26PDD64 pKa = 3.39NDD66 pKa = 4.11GSALTLPSWHH76 pKa = 6.37YY77 pKa = 10.57SCKK80 pKa = 9.35EE81 pKa = 3.76SCMGNHH87 pKa = 7.5LKK89 pKa = 10.45RR90 pKa = 11.84VVNITGARR98 pKa = 11.84WNYY101 pKa = 9.61VGISIPVFKK110 pKa = 10.57IVTNEE115 pKa = 3.44VCYY118 pKa = 10.13TSHH121 pKa = 6.31EE122 pKa = 4.47NVWGYY127 pKa = 10.68CSQYY131 pKa = 10.47QISRR135 pKa = 11.84PVATQKK141 pKa = 11.16SDD143 pKa = 3.37VSCITSSMWDD153 pKa = 3.41NDD155 pKa = 3.07KK156 pKa = 11.61SPIGSLYY163 pKa = 10.87NIVNSNEE170 pKa = 3.81AEE172 pKa = 3.8CDD174 pKa = 3.73YY175 pKa = 11.12FSDD178 pKa = 3.76ITDD181 pKa = 3.63CNRR184 pKa = 11.84DD185 pKa = 3.33YY186 pKa = 11.32QIFKK190 pKa = 10.84RR191 pKa = 11.84EE192 pKa = 3.58GKK194 pKa = 9.96LIKK197 pKa = 10.51RR198 pKa = 11.84SDD200 pKa = 3.86DD201 pKa = 3.64SPLEE205 pKa = 3.93LSIVTDD211 pKa = 5.22GIRR214 pKa = 11.84TDD216 pKa = 3.53PASEE220 pKa = 4.16YY221 pKa = 11.06LSLDD225 pKa = 3.46DD226 pKa = 6.25VSWFWKK232 pKa = 10.42LPNNDD237 pKa = 3.49MSPPCGWEE245 pKa = 3.82KK246 pKa = 9.02TQKK249 pKa = 10.45LSCSYY254 pKa = 11.08TDD256 pKa = 3.14TTDD259 pKa = 3.45VIKK262 pKa = 10.79CNSIGYY268 pKa = 7.22TYY270 pKa = 10.32NIQGISKK277 pKa = 10.5KK278 pKa = 7.87STCAGNIYY286 pKa = 9.28DD287 pKa = 4.27TDD289 pKa = 4.02GPFPFFYY296 pKa = 10.62DD297 pKa = 3.34AEE299 pKa = 4.27EE300 pKa = 5.11ALMSTDD306 pKa = 4.04DD307 pKa = 3.6ACGKK311 pKa = 10.14AKK313 pKa = 10.02QGKK316 pKa = 8.29PDD318 pKa = 3.65ADD320 pKa = 3.12IAFIEE325 pKa = 4.67GVNRR329 pKa = 11.84AFEE332 pKa = 4.64DD333 pKa = 4.64LEE335 pKa = 4.26LTYY338 pKa = 10.97CSATCDD344 pKa = 3.17LFARR348 pKa = 11.84QGTPNEE354 pKa = 4.09DD355 pKa = 3.21HH356 pKa = 7.34VLDD359 pKa = 4.27TPIGTWRR366 pKa = 11.84YY367 pKa = 8.61VMRR370 pKa = 11.84DD371 pKa = 3.4NLDD374 pKa = 3.81PALVPCLPTSNWTISDD390 pKa = 3.67PTTICHH396 pKa = 6.15GKK398 pKa = 9.54DD399 pKa = 3.11HH400 pKa = 6.74ILVVDD405 pKa = 4.28TATGHH410 pKa = 5.81SGSWDD415 pKa = 3.45TKK417 pKa = 9.56KK418 pKa = 10.91DD419 pKa = 3.73YY420 pKa = 10.83IITGEE425 pKa = 4.15VCNTNNDD432 pKa = 3.72EE433 pKa = 4.72MGDD436 pKa = 4.08DD437 pKa = 3.97YY438 pKa = 11.86DD439 pKa = 4.55GMRR442 pKa = 11.84DD443 pKa = 3.3KK444 pKa = 10.72ILRR447 pKa = 11.84GEE449 pKa = 4.18TIEE452 pKa = 4.07IKK454 pKa = 10.32FWTGDD459 pKa = 4.02IIRR462 pKa = 11.84MAPPYY467 pKa = 10.44DD468 pKa = 3.47NPEE471 pKa = 4.17WIKK474 pKa = 11.11GSVLFRR480 pKa = 11.84QNPGWFSSVEE490 pKa = 3.89LNKK493 pKa = 11.05DD494 pKa = 3.62MIHH497 pKa = 5.89TRR499 pKa = 11.84DD500 pKa = 4.11NITDD504 pKa = 4.52LLTVMVQNATAEE516 pKa = 4.21VMYY519 pKa = 10.73KK520 pKa = 10.44RR521 pKa = 11.84LDD523 pKa = 3.78PKK525 pKa = 9.91TMKK528 pKa = 10.29HH529 pKa = 5.54ILFAEE534 pKa = 4.28IVDD537 pKa = 4.07GVGNVSGKK545 pKa = 10.24ISGFLTGLFGGFTKK559 pKa = 10.66AVIIVASLAICYY571 pKa = 9.43IVLSVLWKK579 pKa = 9.37VRR581 pKa = 11.84LVASIFNSAKK591 pKa = 9.73KK592 pKa = 10.11KK593 pKa = 9.69RR594 pKa = 11.84VRR596 pKa = 11.84ISDD599 pKa = 3.47ILDD602 pKa = 3.8EE603 pKa = 4.55EE604 pKa = 4.4PHH606 pKa = 7.09RR607 pKa = 11.84IQQSRR612 pKa = 11.84PTLSRR617 pKa = 11.84KK618 pKa = 9.62KK619 pKa = 9.2KK620 pKa = 7.5TRR622 pKa = 11.84EE623 pKa = 4.0SIQMLLNDD631 pKa = 3.71II632 pKa = 4.81
MM1 pKa = 7.07SHH3 pKa = 6.53IMNLLVISFVLAGSSWSLLGYY24 pKa = 10.25QDD26 pKa = 4.9DD27 pKa = 4.41FSSKK31 pKa = 10.19RR32 pKa = 11.84SGALASNPTYY42 pKa = 10.73NLPQDD47 pKa = 3.27KK48 pKa = 10.87GYY50 pKa = 10.71GRR52 pKa = 11.84DD53 pKa = 3.31MYY55 pKa = 10.5QPYY58 pKa = 9.85YY59 pKa = 10.01ICEE62 pKa = 4.26PDD64 pKa = 3.39NDD66 pKa = 4.11GSALTLPSWHH76 pKa = 6.37YY77 pKa = 10.57SCKK80 pKa = 9.35EE81 pKa = 3.76SCMGNHH87 pKa = 7.5LKK89 pKa = 10.45RR90 pKa = 11.84VVNITGARR98 pKa = 11.84WNYY101 pKa = 9.61VGISIPVFKK110 pKa = 10.57IVTNEE115 pKa = 3.44VCYY118 pKa = 10.13TSHH121 pKa = 6.31EE122 pKa = 4.47NVWGYY127 pKa = 10.68CSQYY131 pKa = 10.47QISRR135 pKa = 11.84PVATQKK141 pKa = 11.16SDD143 pKa = 3.37VSCITSSMWDD153 pKa = 3.41NDD155 pKa = 3.07KK156 pKa = 11.61SPIGSLYY163 pKa = 10.87NIVNSNEE170 pKa = 3.81AEE172 pKa = 3.8CDD174 pKa = 3.73YY175 pKa = 11.12FSDD178 pKa = 3.76ITDD181 pKa = 3.63CNRR184 pKa = 11.84DD185 pKa = 3.33YY186 pKa = 11.32QIFKK190 pKa = 10.84RR191 pKa = 11.84EE192 pKa = 3.58GKK194 pKa = 9.96LIKK197 pKa = 10.51RR198 pKa = 11.84SDD200 pKa = 3.86DD201 pKa = 3.64SPLEE205 pKa = 3.93LSIVTDD211 pKa = 5.22GIRR214 pKa = 11.84TDD216 pKa = 3.53PASEE220 pKa = 4.16YY221 pKa = 11.06LSLDD225 pKa = 3.46DD226 pKa = 6.25VSWFWKK232 pKa = 10.42LPNNDD237 pKa = 3.49MSPPCGWEE245 pKa = 3.82KK246 pKa = 9.02TQKK249 pKa = 10.45LSCSYY254 pKa = 11.08TDD256 pKa = 3.14TTDD259 pKa = 3.45VIKK262 pKa = 10.79CNSIGYY268 pKa = 7.22TYY270 pKa = 10.32NIQGISKK277 pKa = 10.5KK278 pKa = 7.87STCAGNIYY286 pKa = 9.28DD287 pKa = 4.27TDD289 pKa = 4.02GPFPFFYY296 pKa = 10.62DD297 pKa = 3.34AEE299 pKa = 4.27EE300 pKa = 5.11ALMSTDD306 pKa = 4.04DD307 pKa = 3.6ACGKK311 pKa = 10.14AKK313 pKa = 10.02QGKK316 pKa = 8.29PDD318 pKa = 3.65ADD320 pKa = 3.12IAFIEE325 pKa = 4.67GVNRR329 pKa = 11.84AFEE332 pKa = 4.64DD333 pKa = 4.64LEE335 pKa = 4.26LTYY338 pKa = 10.97CSATCDD344 pKa = 3.17LFARR348 pKa = 11.84QGTPNEE354 pKa = 4.09DD355 pKa = 3.21HH356 pKa = 7.34VLDD359 pKa = 4.27TPIGTWRR366 pKa = 11.84YY367 pKa = 8.61VMRR370 pKa = 11.84DD371 pKa = 3.4NLDD374 pKa = 3.81PALVPCLPTSNWTISDD390 pKa = 3.67PTTICHH396 pKa = 6.15GKK398 pKa = 9.54DD399 pKa = 3.11HH400 pKa = 6.74ILVVDD405 pKa = 4.28TATGHH410 pKa = 5.81SGSWDD415 pKa = 3.45TKK417 pKa = 9.56KK418 pKa = 10.91DD419 pKa = 3.73YY420 pKa = 10.83IITGEE425 pKa = 4.15VCNTNNDD432 pKa = 3.72EE433 pKa = 4.72MGDD436 pKa = 4.08DD437 pKa = 3.97YY438 pKa = 11.86DD439 pKa = 4.55GMRR442 pKa = 11.84DD443 pKa = 3.3KK444 pKa = 10.72ILRR447 pKa = 11.84GEE449 pKa = 4.18TIEE452 pKa = 4.07IKK454 pKa = 10.32FWTGDD459 pKa = 4.02IIRR462 pKa = 11.84MAPPYY467 pKa = 10.44DD468 pKa = 3.47NPEE471 pKa = 4.17WIKK474 pKa = 11.11GSVLFRR480 pKa = 11.84QNPGWFSSVEE490 pKa = 3.89LNKK493 pKa = 11.05DD494 pKa = 3.62MIHH497 pKa = 5.89TRR499 pKa = 11.84DD500 pKa = 4.11NITDD504 pKa = 4.52LLTVMVQNATAEE516 pKa = 4.21VMYY519 pKa = 10.73KK520 pKa = 10.44RR521 pKa = 11.84LDD523 pKa = 3.78PKK525 pKa = 9.91TMKK528 pKa = 10.29HH529 pKa = 5.54ILFAEE534 pKa = 4.28IVDD537 pKa = 4.07GVGNVSGKK545 pKa = 10.24ISGFLTGLFGGFTKK559 pKa = 10.66AVIIVASLAICYY571 pKa = 9.43IVLSVLWKK579 pKa = 9.37VRR581 pKa = 11.84LVASIFNSAKK591 pKa = 9.73KK592 pKa = 10.11KK593 pKa = 9.69RR594 pKa = 11.84VRR596 pKa = 11.84ISDD599 pKa = 3.47ILDD602 pKa = 3.8EE603 pKa = 4.55EE604 pKa = 4.4PHH606 pKa = 7.09RR607 pKa = 11.84IQQSRR612 pKa = 11.84PTLSRR617 pKa = 11.84KK618 pKa = 9.62KK619 pKa = 9.2KK620 pKa = 7.5TRR622 pKa = 11.84EE623 pKa = 4.0SIQMLLNDD631 pKa = 3.71II632 pKa = 4.81
Molecular weight: 71.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q89914|MVP_SYNV Movement protein OS=Sonchus yellow net virus OX=11307 GN=sc4 PE=1 SV=1
MM1 pKa = 7.67EE2 pKa = 5.27GLSSKK7 pKa = 10.05AQTMGRR13 pKa = 11.84EE14 pKa = 4.04DD15 pKa = 4.24DD16 pKa = 3.82NRR18 pKa = 11.84SSKK21 pKa = 10.44MKK23 pKa = 10.02VFHH26 pKa = 6.86SEE28 pKa = 3.48LVYY31 pKa = 11.11GDD33 pKa = 3.67NHH35 pKa = 6.7NISIKK40 pKa = 10.57KK41 pKa = 10.05ADD43 pKa = 3.66LTGQHH48 pKa = 6.64KK49 pKa = 10.18MMLLLSSALRR59 pKa = 11.84IGSVHH64 pKa = 6.26MDD66 pKa = 3.06VSRR69 pKa = 11.84ILVKK73 pKa = 9.07WCPYY77 pKa = 8.06ITPNMNTTIGITIKK91 pKa = 10.82NNHH94 pKa = 6.81HH95 pKa = 7.44DD96 pKa = 4.15DD97 pKa = 3.51MSNINDD103 pKa = 3.35MSTYY107 pKa = 10.3ISVKK111 pKa = 10.25GKK113 pKa = 9.67MSEE116 pKa = 3.82ALQITWHH123 pKa = 6.4PASTLVYY130 pKa = 9.32KK131 pKa = 10.72KK132 pKa = 10.77GMSCIFPWVVDD143 pKa = 3.42VDD145 pKa = 3.94TGSTEE150 pKa = 4.06QEE152 pKa = 4.18SGSPALGEE160 pKa = 4.0IKK162 pKa = 9.83IWCYY166 pKa = 11.17FKK168 pKa = 10.32MQYY171 pKa = 9.97HH172 pKa = 6.23KK173 pKa = 10.69PSTRR177 pKa = 11.84HH178 pKa = 4.61IARR181 pKa = 11.84AEE183 pKa = 3.79IAPSIEE189 pKa = 3.69WGNTNFPYY197 pKa = 10.35YY198 pKa = 10.84VPFAMIRR205 pKa = 11.84RR206 pKa = 11.84ARR208 pKa = 11.84GIRR211 pKa = 11.84PLDD214 pKa = 3.42VFSTNQYY221 pKa = 11.65SMFLEE226 pKa = 4.45DD227 pKa = 3.7VIKK230 pKa = 10.74HH231 pKa = 5.65VGTDD235 pKa = 3.64SIKK238 pKa = 10.75EE239 pKa = 3.98SDD241 pKa = 3.74IVPIMSTMSQEE252 pKa = 4.03DD253 pKa = 3.39MMMINEE259 pKa = 4.15KK260 pKa = 10.91NKK262 pKa = 8.88TCLLKK267 pKa = 10.78RR268 pKa = 11.84GGSYY272 pKa = 10.41CSCKK276 pKa = 10.46DD277 pKa = 3.37VIEE280 pKa = 4.33NVVKK284 pKa = 10.18EE285 pKa = 4.05INMNRR290 pKa = 11.84DD291 pKa = 2.95RR292 pKa = 11.84KK293 pKa = 9.87YY294 pKa = 11.23DD295 pKa = 3.49NHH297 pKa = 6.56GLLLSGYY304 pKa = 9.27IAGSTSGRR312 pKa = 11.84FQTVPMLSDD321 pKa = 3.18ISYY324 pKa = 11.01
MM1 pKa = 7.67EE2 pKa = 5.27GLSSKK7 pKa = 10.05AQTMGRR13 pKa = 11.84EE14 pKa = 4.04DD15 pKa = 4.24DD16 pKa = 3.82NRR18 pKa = 11.84SSKK21 pKa = 10.44MKK23 pKa = 10.02VFHH26 pKa = 6.86SEE28 pKa = 3.48LVYY31 pKa = 11.11GDD33 pKa = 3.67NHH35 pKa = 6.7NISIKK40 pKa = 10.57KK41 pKa = 10.05ADD43 pKa = 3.66LTGQHH48 pKa = 6.64KK49 pKa = 10.18MMLLLSSALRR59 pKa = 11.84IGSVHH64 pKa = 6.26MDD66 pKa = 3.06VSRR69 pKa = 11.84ILVKK73 pKa = 9.07WCPYY77 pKa = 8.06ITPNMNTTIGITIKK91 pKa = 10.82NNHH94 pKa = 6.81HH95 pKa = 7.44DD96 pKa = 4.15DD97 pKa = 3.51MSNINDD103 pKa = 3.35MSTYY107 pKa = 10.3ISVKK111 pKa = 10.25GKK113 pKa = 9.67MSEE116 pKa = 3.82ALQITWHH123 pKa = 6.4PASTLVYY130 pKa = 9.32KK131 pKa = 10.72KK132 pKa = 10.77GMSCIFPWVVDD143 pKa = 3.42VDD145 pKa = 3.94TGSTEE150 pKa = 4.06QEE152 pKa = 4.18SGSPALGEE160 pKa = 4.0IKK162 pKa = 9.83IWCYY166 pKa = 11.17FKK168 pKa = 10.32MQYY171 pKa = 9.97HH172 pKa = 6.23KK173 pKa = 10.69PSTRR177 pKa = 11.84HH178 pKa = 4.61IARR181 pKa = 11.84AEE183 pKa = 3.79IAPSIEE189 pKa = 3.69WGNTNFPYY197 pKa = 10.35YY198 pKa = 10.84VPFAMIRR205 pKa = 11.84RR206 pKa = 11.84ARR208 pKa = 11.84GIRR211 pKa = 11.84PLDD214 pKa = 3.42VFSTNQYY221 pKa = 11.65SMFLEE226 pKa = 4.45DD227 pKa = 3.7VIKK230 pKa = 10.74HH231 pKa = 5.65VGTDD235 pKa = 3.64SIKK238 pKa = 10.75EE239 pKa = 3.98SDD241 pKa = 3.74IVPIMSTMSQEE252 pKa = 4.03DD253 pKa = 3.39MMMINEE259 pKa = 4.15KK260 pKa = 10.91NKK262 pKa = 8.88TCLLKK267 pKa = 10.78RR268 pKa = 11.84GGSYY272 pKa = 10.41CSCKK276 pKa = 10.46DD277 pKa = 3.37VIEE280 pKa = 4.33NVVKK284 pKa = 10.18EE285 pKa = 4.05INMNRR290 pKa = 11.84DD291 pKa = 2.95RR292 pKa = 11.84KK293 pKa = 9.87YY294 pKa = 11.23DD295 pKa = 3.49NHH297 pKa = 6.56GLLLSGYY304 pKa = 9.27IAGSTSGRR312 pKa = 11.84FQTVPMLSDD321 pKa = 3.18ISYY324 pKa = 11.01
Molecular weight: 36.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4178 |
286 |
2116 |
696.3 |
78.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.05 ± 0.695 | 2.106 ± 0.243 |
6.726 ± 0.433 | 5.05 ± 0.354 |
3.255 ± 0.316 | 5.505 ± 0.454 |
2.8 ± 0.308 | 8.042 ± 0.403 |
6.462 ± 0.322 | 8.664 ± 0.991 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.303 ± 0.429 | 5.218 ± 0.119 |
3.71 ± 0.346 | 2.824 ± 0.12 |
4.859 ± 0.556 | 9.023 ± 0.253 |
6.415 ± 0.261 | 5.385 ± 0.339 |
1.316 ± 0.24 | 4.284 ± 0.118 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
