
Eragrostis streak virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
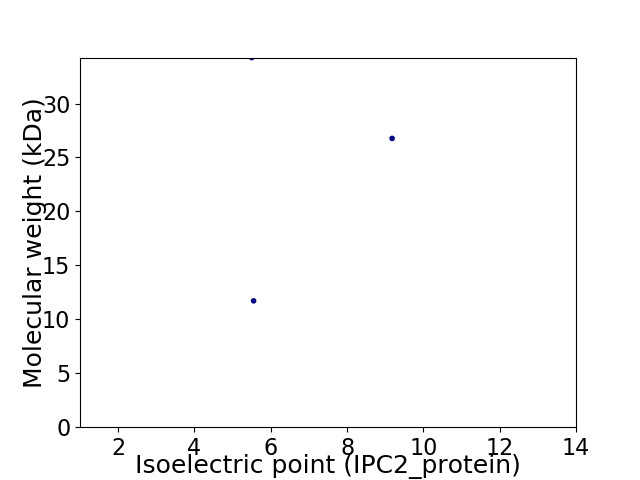
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
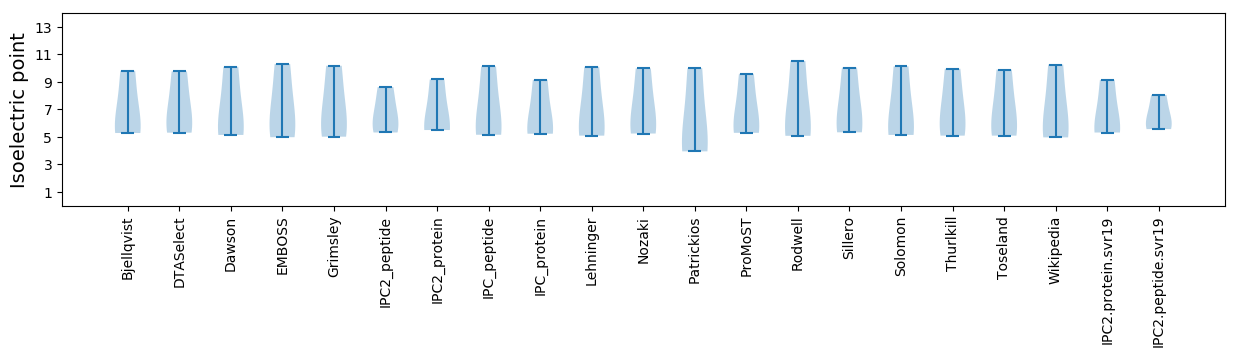
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B0Z3X7|B0Z3X7_9GEMI Capsid protein OS=Eragrostis streak virus OX=496807 PE=3 SV=1
MM1 pKa = 7.71EE2 pKa = 4.89SFEE5 pKa = 4.43GAGSFVPQVALPRR18 pKa = 11.84VPPAAPSVPSLPWSSVGEE36 pKa = 3.59IAIFIFVAVLALYY49 pKa = 9.25LLWIWVVRR57 pKa = 11.84DD58 pKa = 3.29CLFVVKK64 pKa = 10.42ARR66 pKa = 11.84QGGSTEE72 pKa = 3.95EE73 pKa = 4.08LQFGPRR79 pKa = 11.84EE80 pKa = 4.13RR81 pKa = 11.84PPVPAGDD88 pKa = 4.07RR89 pKa = 11.84PPLPPAVSVATTSEE103 pKa = 3.95TRR105 pKa = 11.84PFSVV109 pKa = 3.54
MM1 pKa = 7.71EE2 pKa = 4.89SFEE5 pKa = 4.43GAGSFVPQVALPRR18 pKa = 11.84VPPAAPSVPSLPWSSVGEE36 pKa = 3.59IAIFIFVAVLALYY49 pKa = 9.25LLWIWVVRR57 pKa = 11.84DD58 pKa = 3.29CLFVVKK64 pKa = 10.42ARR66 pKa = 11.84QGGSTEE72 pKa = 3.95EE73 pKa = 4.08LQFGPRR79 pKa = 11.84EE80 pKa = 4.13RR81 pKa = 11.84PPVPAGDD88 pKa = 4.07RR89 pKa = 11.84PPLPPAVSVATTSEE103 pKa = 3.95TRR105 pKa = 11.84PFSVV109 pKa = 3.54
Molecular weight: 11.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B0Z3X8|B0Z3X8_9GEMI Replication-associated protein OS=Eragrostis streak virus OX=496807 PE=3 SV=1
MM1 pKa = 7.89PLTAGKK7 pKa = 10.09RR8 pKa = 11.84KK9 pKa = 9.88RR10 pKa = 11.84SGDD13 pKa = 3.37ASSSKK18 pKa = 10.07VARR21 pKa = 11.84PKK23 pKa = 10.51LGRR26 pKa = 11.84SSAARR31 pKa = 11.84AGKK34 pKa = 9.18PSTRR38 pKa = 11.84ARR40 pKa = 11.84PSLQIQTLQAAGQSMISVPSGGVCDD65 pKa = 4.55LLGSFARR72 pKa = 11.84GSDD75 pKa = 2.95EE76 pKa = 4.47GNRR79 pKa = 11.84HH80 pKa = 4.54TNEE83 pKa = 3.6TVTYY87 pKa = 9.82KK88 pKa = 10.8VALDD92 pKa = 3.36YY93 pKa = 11.49HH94 pKa = 6.46FVATAGACKK103 pKa = 10.04YY104 pKa = 10.58SSIGTGVVWLVYY116 pKa = 10.04DD117 pKa = 4.3AQPQGNCPQVKK128 pKa = 10.28DD129 pKa = 3.98IFPHH133 pKa = 6.65PDD135 pKa = 2.96SLSAFPYY142 pKa = 6.97TWKK145 pKa = 10.5VGRR148 pKa = 11.84EE149 pKa = 3.83VCHH152 pKa = 6.21RR153 pKa = 11.84FVVKK157 pKa = 10.4RR158 pKa = 11.84RR159 pKa = 11.84WLFTMEE165 pKa = 4.01TNGRR169 pKa = 11.84IGSDD173 pKa = 3.29TPGPTSCWPPCRR185 pKa = 11.84KK186 pKa = 9.9NIYY189 pKa = 7.73FHH191 pKa = 7.04KK192 pKa = 10.44FEE194 pKa = 5.89AGLGVKK200 pKa = 7.97TEE202 pKa = 4.16WKK204 pKa = 8.16NTTGGDD210 pKa = 3.17VGDD213 pKa = 3.87IKK215 pKa = 10.91KK216 pKa = 10.21GALYY220 pKa = 10.14IVIAPGNGLEE230 pKa = 4.27FTVHH234 pKa = 5.73GNARR238 pKa = 11.84LYY240 pKa = 10.33FKK242 pKa = 10.94SVGNQQ247 pKa = 2.8
MM1 pKa = 7.89PLTAGKK7 pKa = 10.09RR8 pKa = 11.84KK9 pKa = 9.88RR10 pKa = 11.84SGDD13 pKa = 3.37ASSSKK18 pKa = 10.07VARR21 pKa = 11.84PKK23 pKa = 10.51LGRR26 pKa = 11.84SSAARR31 pKa = 11.84AGKK34 pKa = 9.18PSTRR38 pKa = 11.84ARR40 pKa = 11.84PSLQIQTLQAAGQSMISVPSGGVCDD65 pKa = 4.55LLGSFARR72 pKa = 11.84GSDD75 pKa = 2.95EE76 pKa = 4.47GNRR79 pKa = 11.84HH80 pKa = 4.54TNEE83 pKa = 3.6TVTYY87 pKa = 9.82KK88 pKa = 10.8VALDD92 pKa = 3.36YY93 pKa = 11.49HH94 pKa = 6.46FVATAGACKK103 pKa = 10.04YY104 pKa = 10.58SSIGTGVVWLVYY116 pKa = 10.04DD117 pKa = 4.3AQPQGNCPQVKK128 pKa = 10.28DD129 pKa = 3.98IFPHH133 pKa = 6.65PDD135 pKa = 2.96SLSAFPYY142 pKa = 6.97TWKK145 pKa = 10.5VGRR148 pKa = 11.84EE149 pKa = 3.83VCHH152 pKa = 6.21RR153 pKa = 11.84FVVKK157 pKa = 10.4RR158 pKa = 11.84RR159 pKa = 11.84WLFTMEE165 pKa = 4.01TNGRR169 pKa = 11.84IGSDD173 pKa = 3.29TPGPTSCWPPCRR185 pKa = 11.84KK186 pKa = 9.9NIYY189 pKa = 7.73FHH191 pKa = 7.04KK192 pKa = 10.44FEE194 pKa = 5.89AGLGVKK200 pKa = 7.97TEE202 pKa = 4.16WKK204 pKa = 8.16NTTGGDD210 pKa = 3.17VGDD213 pKa = 3.87IKK215 pKa = 10.91KK216 pKa = 10.21GALYY220 pKa = 10.14IVIAPGNGLEE230 pKa = 4.27FTVHH234 pKa = 5.73GNARR238 pKa = 11.84LYY240 pKa = 10.33FKK242 pKa = 10.94SVGNQQ247 pKa = 2.8
Molecular weight: 26.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
656 |
109 |
300 |
218.7 |
24.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.622 ± 0.974 | 1.372 ± 0.449 |
3.963 ± 0.736 | 5.335 ± 1.062 |
4.116 ± 0.801 | 6.86 ± 1.999 |
2.591 ± 0.944 | 4.268 ± 0.415 |
4.726 ± 1.329 | 6.86 ± 0.58 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.22 ± 0.107 | 2.896 ± 0.944 |
7.774 ± 2.253 | 4.573 ± 1.041 |
5.64 ± 0.551 | 9.299 ± 0.569 |
7.622 ± 1.553 | 7.47 ± 2.257 |
2.439 ± 0.18 | 3.354 ± 0.879 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
