
Mycoplasma phocirhinis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasmopsis
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins
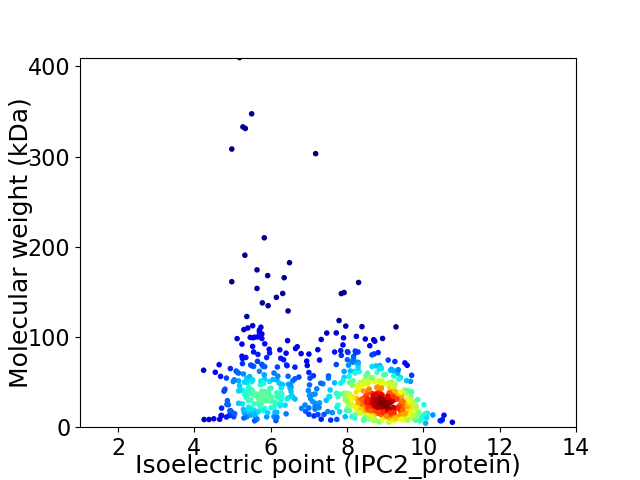
Virtual 2D-PAGE plot for 676 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
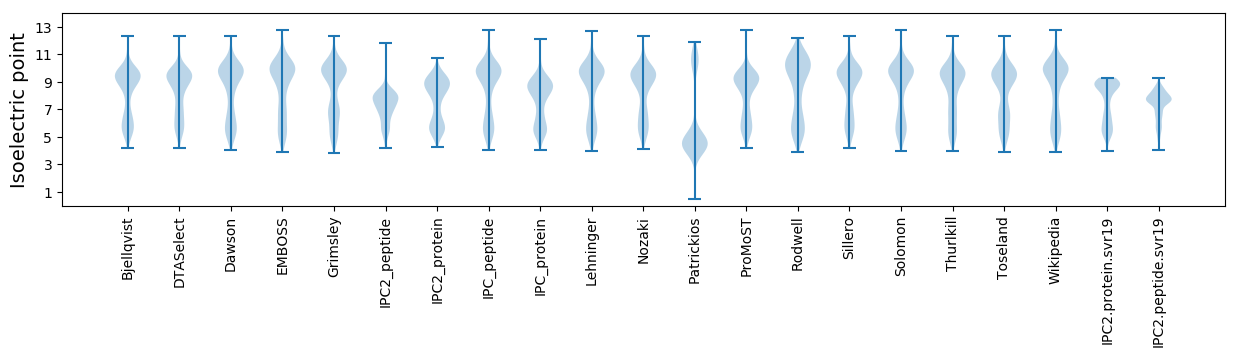
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P6MTV0|A0A4P6MTV0_9MOLU HAD family phosphatase OS=Mycoplasma phocirhinis OX=142650 GN=EG856_02740 PE=4 SV=1
MM1 pKa = 7.33NKK3 pKa = 10.21DD4 pKa = 3.49KK5 pKa = 11.31NILLNTEE12 pKa = 3.81YY13 pKa = 10.19LTRR16 pKa = 11.84LEE18 pKa = 3.87QNEE21 pKa = 4.26IYY23 pKa = 10.38KK24 pKa = 10.78AILNYY29 pKa = 10.05DD30 pKa = 3.85SKK32 pKa = 11.53LSAIVQATKK41 pKa = 10.5KK42 pKa = 7.77QTISQTDD49 pKa = 3.48YY50 pKa = 11.25FRR52 pKa = 11.84IKK54 pKa = 11.02SNFNQVEE61 pKa = 4.06DD62 pKa = 5.4LIDD65 pKa = 4.48DD66 pKa = 4.96LLTISRR72 pKa = 11.84FNYY75 pKa = 9.99LEE77 pKa = 4.12EE78 pKa = 4.28VKK80 pKa = 10.55SLYY83 pKa = 10.92ASIVNNFNLLFTKK96 pKa = 10.35YY97 pKa = 10.58VIDD100 pKa = 3.84YY101 pKa = 7.46TASDD105 pKa = 3.82SPDD108 pKa = 2.57TRR110 pKa = 11.84AYY112 pKa = 10.41HH113 pKa = 7.12KK114 pKa = 10.78LDD116 pKa = 3.43TEE118 pKa = 4.4QQFSLTSYY126 pKa = 8.18LTSFVIVEE134 pKa = 4.16DD135 pKa = 3.54KK136 pKa = 10.95FVIFVAVDD144 pKa = 3.68KK145 pKa = 10.54YY146 pKa = 10.86QYY148 pKa = 10.32PILTEE153 pKa = 4.49FVCSEE158 pKa = 4.25TDD160 pKa = 4.1FSCPADD166 pKa = 3.91CEE168 pKa = 4.33KK169 pKa = 10.88SCCLVQVQAVEE180 pKa = 4.04NSDD183 pKa = 3.34MAEE186 pKa = 4.04TQNITEE192 pKa = 4.62EE193 pKa = 5.02DD194 pKa = 4.33YY195 pKa = 11.51SCPTDD200 pKa = 4.27CEE202 pKa = 4.32KK203 pKa = 10.41TCCVTEE209 pKa = 4.16VQAVEE214 pKa = 4.07NSDD217 pKa = 3.58IEE219 pKa = 4.64TNTNEE224 pKa = 4.12NEE226 pKa = 4.15TTPVEE231 pKa = 4.73FIEE234 pKa = 4.09EE235 pKa = 4.12TKK237 pKa = 10.62INEE240 pKa = 3.8EE241 pKa = 3.94NLILDD246 pKa = 3.96HH247 pKa = 7.37SDD249 pKa = 2.97VLTQTEE255 pKa = 4.49FNQYY259 pKa = 10.56LDD261 pKa = 3.78LAHH264 pKa = 5.79YY265 pKa = 8.38TSLPEE270 pKa = 5.1KK271 pKa = 10.57DD272 pKa = 4.86LLDD275 pKa = 4.28ALTKK279 pKa = 10.52YY280 pKa = 8.59SHH282 pKa = 7.03KK283 pKa = 10.66LEE285 pKa = 5.02KK286 pKa = 10.53INTNFKK292 pKa = 10.39RR293 pKa = 11.84QTVSQADD300 pKa = 3.75YY301 pKa = 11.05FRR303 pKa = 11.84TKK305 pKa = 10.78QNLNDD310 pKa = 3.63VQEE313 pKa = 4.56LVDD316 pKa = 5.76DD317 pKa = 5.04LLTHH321 pKa = 7.37DD322 pKa = 4.16SYY324 pKa = 11.98SYY326 pKa = 10.52KK327 pKa = 10.42QEE329 pKa = 3.99TQLVYY334 pKa = 11.28DD335 pKa = 3.9DD336 pKa = 4.03VNRR339 pKa = 11.84NFDD342 pKa = 4.13NILNNLVIDD351 pKa = 3.78YY352 pKa = 7.56TAADD356 pKa = 3.84TPDD359 pKa = 3.59NNGLALLNINEE370 pKa = 4.32NDD372 pKa = 3.3FLNYY376 pKa = 7.14QTSFILHH383 pKa = 5.92EE384 pKa = 4.5NKK386 pKa = 9.78QVVFNVANEE395 pKa = 3.82YY396 pKa = 10.55SYY398 pKa = 10.94PILSVQNLEE407 pKa = 4.27TNSIEE412 pKa = 4.41DD413 pKa = 3.48SSEE416 pKa = 4.23HH417 pKa = 5.61EE418 pKa = 3.79QHH420 pKa = 6.71LMNEE424 pKa = 4.26MLVEE428 pKa = 4.01NSVVEE433 pKa = 4.4PIMVDD438 pKa = 3.3EE439 pKa = 4.68EE440 pKa = 4.64VIEE443 pKa = 4.58HH444 pKa = 6.31QDD446 pKa = 2.96KK447 pKa = 10.44TEE449 pKa = 3.88NNEE452 pKa = 3.68FVEE455 pKa = 5.3FVQSEE460 pKa = 4.41EE461 pKa = 4.2VQNSCNKK468 pKa = 9.89EE469 pKa = 3.83EE470 pKa = 4.91CEE472 pKa = 4.52TNCEE476 pKa = 4.15CASEE480 pKa = 4.16CDD482 pKa = 3.38ITVHH486 pKa = 5.08EE487 pKa = 4.44TQTFVVDD494 pKa = 3.84SNKK497 pKa = 9.93KK498 pKa = 9.66LKK500 pKa = 9.85PSRR503 pKa = 11.84YY504 pKa = 8.96LSKK507 pKa = 10.5PKK509 pKa = 10.38KK510 pKa = 9.42FWWILSLVVVSLILVVLLILVGLRR534 pKa = 11.84AYY536 pKa = 9.85EE537 pKa = 4.13IINNIDD543 pKa = 2.97IFF545 pKa = 4.1
MM1 pKa = 7.33NKK3 pKa = 10.21DD4 pKa = 3.49KK5 pKa = 11.31NILLNTEE12 pKa = 3.81YY13 pKa = 10.19LTRR16 pKa = 11.84LEE18 pKa = 3.87QNEE21 pKa = 4.26IYY23 pKa = 10.38KK24 pKa = 10.78AILNYY29 pKa = 10.05DD30 pKa = 3.85SKK32 pKa = 11.53LSAIVQATKK41 pKa = 10.5KK42 pKa = 7.77QTISQTDD49 pKa = 3.48YY50 pKa = 11.25FRR52 pKa = 11.84IKK54 pKa = 11.02SNFNQVEE61 pKa = 4.06DD62 pKa = 5.4LIDD65 pKa = 4.48DD66 pKa = 4.96LLTISRR72 pKa = 11.84FNYY75 pKa = 9.99LEE77 pKa = 4.12EE78 pKa = 4.28VKK80 pKa = 10.55SLYY83 pKa = 10.92ASIVNNFNLLFTKK96 pKa = 10.35YY97 pKa = 10.58VIDD100 pKa = 3.84YY101 pKa = 7.46TASDD105 pKa = 3.82SPDD108 pKa = 2.57TRR110 pKa = 11.84AYY112 pKa = 10.41HH113 pKa = 7.12KK114 pKa = 10.78LDD116 pKa = 3.43TEE118 pKa = 4.4QQFSLTSYY126 pKa = 8.18LTSFVIVEE134 pKa = 4.16DD135 pKa = 3.54KK136 pKa = 10.95FVIFVAVDD144 pKa = 3.68KK145 pKa = 10.54YY146 pKa = 10.86QYY148 pKa = 10.32PILTEE153 pKa = 4.49FVCSEE158 pKa = 4.25TDD160 pKa = 4.1FSCPADD166 pKa = 3.91CEE168 pKa = 4.33KK169 pKa = 10.88SCCLVQVQAVEE180 pKa = 4.04NSDD183 pKa = 3.34MAEE186 pKa = 4.04TQNITEE192 pKa = 4.62EE193 pKa = 5.02DD194 pKa = 4.33YY195 pKa = 11.51SCPTDD200 pKa = 4.27CEE202 pKa = 4.32KK203 pKa = 10.41TCCVTEE209 pKa = 4.16VQAVEE214 pKa = 4.07NSDD217 pKa = 3.58IEE219 pKa = 4.64TNTNEE224 pKa = 4.12NEE226 pKa = 4.15TTPVEE231 pKa = 4.73FIEE234 pKa = 4.09EE235 pKa = 4.12TKK237 pKa = 10.62INEE240 pKa = 3.8EE241 pKa = 3.94NLILDD246 pKa = 3.96HH247 pKa = 7.37SDD249 pKa = 2.97VLTQTEE255 pKa = 4.49FNQYY259 pKa = 10.56LDD261 pKa = 3.78LAHH264 pKa = 5.79YY265 pKa = 8.38TSLPEE270 pKa = 5.1KK271 pKa = 10.57DD272 pKa = 4.86LLDD275 pKa = 4.28ALTKK279 pKa = 10.52YY280 pKa = 8.59SHH282 pKa = 7.03KK283 pKa = 10.66LEE285 pKa = 5.02KK286 pKa = 10.53INTNFKK292 pKa = 10.39RR293 pKa = 11.84QTVSQADD300 pKa = 3.75YY301 pKa = 11.05FRR303 pKa = 11.84TKK305 pKa = 10.78QNLNDD310 pKa = 3.63VQEE313 pKa = 4.56LVDD316 pKa = 5.76DD317 pKa = 5.04LLTHH321 pKa = 7.37DD322 pKa = 4.16SYY324 pKa = 11.98SYY326 pKa = 10.52KK327 pKa = 10.42QEE329 pKa = 3.99TQLVYY334 pKa = 11.28DD335 pKa = 3.9DD336 pKa = 4.03VNRR339 pKa = 11.84NFDD342 pKa = 4.13NILNNLVIDD351 pKa = 3.78YY352 pKa = 7.56TAADD356 pKa = 3.84TPDD359 pKa = 3.59NNGLALLNINEE370 pKa = 4.32NDD372 pKa = 3.3FLNYY376 pKa = 7.14QTSFILHH383 pKa = 5.92EE384 pKa = 4.5NKK386 pKa = 9.78QVVFNVANEE395 pKa = 3.82YY396 pKa = 10.55SYY398 pKa = 10.94PILSVQNLEE407 pKa = 4.27TNSIEE412 pKa = 4.41DD413 pKa = 3.48SSEE416 pKa = 4.23HH417 pKa = 5.61EE418 pKa = 3.79QHH420 pKa = 6.71LMNEE424 pKa = 4.26MLVEE428 pKa = 4.01NSVVEE433 pKa = 4.4PIMVDD438 pKa = 3.3EE439 pKa = 4.68EE440 pKa = 4.64VIEE443 pKa = 4.58HH444 pKa = 6.31QDD446 pKa = 2.96KK447 pKa = 10.44TEE449 pKa = 3.88NNEE452 pKa = 3.68FVEE455 pKa = 5.3FVQSEE460 pKa = 4.41EE461 pKa = 4.2VQNSCNKK468 pKa = 9.89EE469 pKa = 3.83EE470 pKa = 4.91CEE472 pKa = 4.52TNCEE476 pKa = 4.15CASEE480 pKa = 4.16CDD482 pKa = 3.38ITVHH486 pKa = 5.08EE487 pKa = 4.44TQTFVVDD494 pKa = 3.84SNKK497 pKa = 9.93KK498 pKa = 9.66LKK500 pKa = 9.85PSRR503 pKa = 11.84YY504 pKa = 8.96LSKK507 pKa = 10.5PKK509 pKa = 10.38KK510 pKa = 9.42FWWILSLVVVSLILVVLLILVGLRR534 pKa = 11.84AYY536 pKa = 9.85EE537 pKa = 4.13IINNIDD543 pKa = 2.97IFF545 pKa = 4.1
Molecular weight: 63.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P6MNY0|A0A4P6MNY0_9MOLU Cation-translocating P-type ATPase OS=Mycoplasma phocirhinis OX=142650 GN=EG856_02490 PE=4 SV=1
MM1 pKa = 7.52ARR3 pKa = 11.84ILNIEE8 pKa = 3.76IPNNKK13 pKa = 8.93RR14 pKa = 11.84VVVSLTYY21 pKa = 10.35IFGIGLTRR29 pKa = 11.84SKK31 pKa = 10.93QILAKK36 pKa = 10.84ANINEE41 pKa = 4.33NIRR44 pKa = 11.84VKK46 pKa = 10.72DD47 pKa = 3.72LSEE50 pKa = 4.16EE51 pKa = 3.84QLSAIRR57 pKa = 11.84EE58 pKa = 4.16VARR61 pKa = 11.84EE62 pKa = 3.89YY63 pKa = 9.51QTEE66 pKa = 4.25GDD68 pKa = 3.62LHH70 pKa = 8.28RR71 pKa = 11.84EE72 pKa = 3.37VSLNIKK78 pKa = 10.05RR79 pKa = 11.84LMEE82 pKa = 4.14IKK84 pKa = 10.07CYY86 pKa = 10.37RR87 pKa = 11.84GMRR90 pKa = 11.84HH91 pKa = 6.14RR92 pKa = 11.84KK93 pKa = 7.99GLPVRR98 pKa = 11.84GQSTKK103 pKa = 10.87SNARR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.46GPRR113 pKa = 11.84KK114 pKa = 7.66TVAGKK119 pKa = 10.41KK120 pKa = 9.46KK121 pKa = 10.5
MM1 pKa = 7.52ARR3 pKa = 11.84ILNIEE8 pKa = 3.76IPNNKK13 pKa = 8.93RR14 pKa = 11.84VVVSLTYY21 pKa = 10.35IFGIGLTRR29 pKa = 11.84SKK31 pKa = 10.93QILAKK36 pKa = 10.84ANINEE41 pKa = 4.33NIRR44 pKa = 11.84VKK46 pKa = 10.72DD47 pKa = 3.72LSEE50 pKa = 4.16EE51 pKa = 3.84QLSAIRR57 pKa = 11.84EE58 pKa = 4.16VARR61 pKa = 11.84EE62 pKa = 3.89YY63 pKa = 9.51QTEE66 pKa = 4.25GDD68 pKa = 3.62LHH70 pKa = 8.28RR71 pKa = 11.84EE72 pKa = 3.37VSLNIKK78 pKa = 10.05RR79 pKa = 11.84LMEE82 pKa = 4.14IKK84 pKa = 10.07CYY86 pKa = 10.37RR87 pKa = 11.84GMRR90 pKa = 11.84HH91 pKa = 6.14RR92 pKa = 11.84KK93 pKa = 7.99GLPVRR98 pKa = 11.84GQSTKK103 pKa = 10.87SNARR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.46GPRR113 pKa = 11.84KK114 pKa = 7.66TVAGKK119 pKa = 10.41KK120 pKa = 9.46KK121 pKa = 10.5
Molecular weight: 13.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
260632 |
37 |
3640 |
385.6 |
44.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.393 ± 0.126 | 0.423 ± 0.024 |
5.418 ± 0.083 | 6.044 ± 0.076 |
5.353 ± 0.084 | 4.358 ± 0.095 |
1.529 ± 0.042 | 9.554 ± 0.121 |
9.683 ± 0.076 | 9.494 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.611 ± 0.049 | 8.962 ± 0.18 |
2.627 ± 0.045 | 4.418 ± 0.079 |
3.007 ± 0.053 | 6.567 ± 0.067 |
5.272 ± 0.063 | 5.217 ± 0.073 |
0.92 ± 0.027 | 4.149 ± 0.061 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
