
Exophiala spinifera
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta;
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
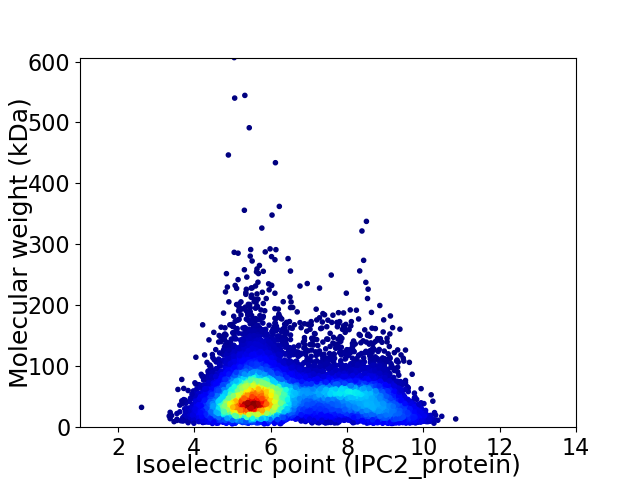
Virtual 2D-PAGE plot for 12047 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D1ZC81|A0A0D1ZC81_9EURO FAD-binding PCMH-type domain-containing protein OS=Exophiala spinifera OX=91928 GN=PV08_11596 PE=3 SV=1
MM1 pKa = 7.77TITGLLLAALAGTAIAQSSDD21 pKa = 3.11LVTVLNSTNGTATFSEE37 pKa = 4.79VLAEE41 pKa = 3.96IPGFLEE47 pKa = 5.01AIAGKK52 pKa = 8.34TNITVLAPNDD62 pKa = 3.55TAIAEE67 pKa = 4.43LLASPAGQALEE78 pKa = 5.39DD79 pKa = 3.98GGADD83 pKa = 3.39YY84 pKa = 11.52ALNLVLYY91 pKa = 9.48HH92 pKa = 6.55VIDD95 pKa = 3.95GGYY98 pKa = 10.98EE99 pKa = 4.02NITDD103 pKa = 3.91YY104 pKa = 11.67AVVPTLLTSSNYY116 pKa = 9.48TNVTGGQHH124 pKa = 6.42IGFYY128 pKa = 10.05WDD130 pKa = 3.85DD131 pKa = 4.44DD132 pKa = 4.21EE133 pKa = 6.65DD134 pKa = 4.23VVGFYY139 pKa = 10.74GGNDD143 pKa = 3.59FAPEE147 pKa = 3.97ASHH150 pKa = 6.48TPIPFDD156 pKa = 3.27QGWIYY161 pKa = 10.77VLNGVLQIPPSMSEE175 pKa = 4.15TVTSGDD181 pKa = 3.62FNGSSFVAALEE192 pKa = 4.08KK193 pKa = 10.88AGLKK197 pKa = 10.5EE198 pKa = 4.25EE199 pKa = 4.41FDD201 pKa = 4.39LLHH204 pKa = 7.07DD205 pKa = 3.63ATYY208 pKa = 9.72FVPVNDD214 pKa = 4.32GFDD217 pKa = 3.48AVEE220 pKa = 4.39DD221 pKa = 4.09ALSKK225 pKa = 11.11LSTEE229 pKa = 4.28EE230 pKa = 3.58LAEE233 pKa = 3.92VLKK236 pKa = 10.24YY237 pKa = 10.42HH238 pKa = 6.08VVAGKK243 pKa = 9.97VWDD246 pKa = 4.12FEE248 pKa = 4.48LLEE251 pKa = 4.52KK252 pKa = 10.73VSQVTTLQGKK262 pKa = 8.65NLTVTQTPDD271 pKa = 3.31EE272 pKa = 4.19EE273 pKa = 4.52WFINQAGIHH282 pKa = 4.82YY283 pKa = 8.6TDD285 pKa = 4.31LAVAGGVVLFIDD297 pKa = 4.08NVLNPLAADD306 pKa = 4.49FAAPRR311 pKa = 11.84NGTDD315 pKa = 4.11GEE317 pKa = 4.41GLPAFTVTNATSTSGTNDD335 pKa = 3.24STSSTASASPSIASATSTGLAAMPLSVGISTLIGGIAVALLFF377 pKa = 4.84
MM1 pKa = 7.77TITGLLLAALAGTAIAQSSDD21 pKa = 3.11LVTVLNSTNGTATFSEE37 pKa = 4.79VLAEE41 pKa = 3.96IPGFLEE47 pKa = 5.01AIAGKK52 pKa = 8.34TNITVLAPNDD62 pKa = 3.55TAIAEE67 pKa = 4.43LLASPAGQALEE78 pKa = 5.39DD79 pKa = 3.98GGADD83 pKa = 3.39YY84 pKa = 11.52ALNLVLYY91 pKa = 9.48HH92 pKa = 6.55VIDD95 pKa = 3.95GGYY98 pKa = 10.98EE99 pKa = 4.02NITDD103 pKa = 3.91YY104 pKa = 11.67AVVPTLLTSSNYY116 pKa = 9.48TNVTGGQHH124 pKa = 6.42IGFYY128 pKa = 10.05WDD130 pKa = 3.85DD131 pKa = 4.44DD132 pKa = 4.21EE133 pKa = 6.65DD134 pKa = 4.23VVGFYY139 pKa = 10.74GGNDD143 pKa = 3.59FAPEE147 pKa = 3.97ASHH150 pKa = 6.48TPIPFDD156 pKa = 3.27QGWIYY161 pKa = 10.77VLNGVLQIPPSMSEE175 pKa = 4.15TVTSGDD181 pKa = 3.62FNGSSFVAALEE192 pKa = 4.08KK193 pKa = 10.88AGLKK197 pKa = 10.5EE198 pKa = 4.25EE199 pKa = 4.41FDD201 pKa = 4.39LLHH204 pKa = 7.07DD205 pKa = 3.63ATYY208 pKa = 9.72FVPVNDD214 pKa = 4.32GFDD217 pKa = 3.48AVEE220 pKa = 4.39DD221 pKa = 4.09ALSKK225 pKa = 11.11LSTEE229 pKa = 4.28EE230 pKa = 3.58LAEE233 pKa = 3.92VLKK236 pKa = 10.24YY237 pKa = 10.42HH238 pKa = 6.08VVAGKK243 pKa = 9.97VWDD246 pKa = 4.12FEE248 pKa = 4.48LLEE251 pKa = 4.52KK252 pKa = 10.73VSQVTTLQGKK262 pKa = 8.65NLTVTQTPDD271 pKa = 3.31EE272 pKa = 4.19EE273 pKa = 4.52WFINQAGIHH282 pKa = 4.82YY283 pKa = 8.6TDD285 pKa = 4.31LAVAGGVVLFIDD297 pKa = 4.08NVLNPLAADD306 pKa = 4.49FAAPRR311 pKa = 11.84NGTDD315 pKa = 4.11GEE317 pKa = 4.41GLPAFTVTNATSTSGTNDD335 pKa = 3.24STSSTASASPSIASATSTGLAAMPLSVGISTLIGGIAVALLFF377 pKa = 4.84
Molecular weight: 39.26 kDa
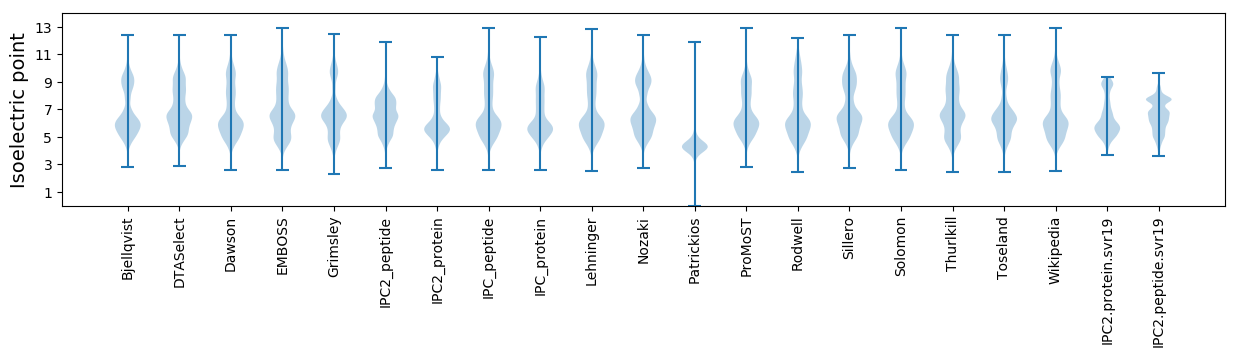
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2BY59|A0A0D2BY59_9EURO T5orf172 domain-containing protein OS=Exophiala spinifera OX=91928 GN=PV08_06305 PE=4 SV=1
MM1 pKa = 7.89PDD3 pKa = 3.61DD4 pKa = 4.49VDD6 pKa = 4.33MKK8 pKa = 9.84STCNVDD14 pKa = 3.13GNQVTKK20 pKa = 11.32NKK22 pKa = 8.86MFSGNWMVVMEE33 pKa = 4.76GLTFAWMRR41 pKa = 11.84TFEE44 pKa = 4.38IVAGPPVTITSTPTATYY61 pKa = 8.99TITSTPRR68 pKa = 11.84ATVTTTSTDD77 pKa = 2.77IYY79 pKa = 10.13STTLSPLTVTVPSSTSTRR97 pKa = 11.84TITTTPKK104 pKa = 10.27QVTVDD109 pKa = 3.54STSTSTRR116 pKa = 11.84TRR118 pKa = 11.84ISRR121 pKa = 11.84TWSATVTTTTVTVSCQPQIVKK142 pKa = 10.09RR143 pKa = 11.84DD144 pKa = 3.75PTCTIRR150 pKa = 11.84PTKK153 pKa = 9.61ATLAAASNTTYY164 pKa = 9.78PAPRR168 pKa = 11.84GRR170 pKa = 11.84FFRR173 pKa = 11.84GAPRR177 pKa = 11.84RR178 pKa = 11.84SRR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84EE183 pKa = 3.72AFVEE187 pKa = 3.94PRR189 pKa = 11.84RR190 pKa = 11.84EE191 pKa = 3.83NQLFEE196 pKa = 5.0RR197 pKa = 11.84GAGTLLL203 pKa = 3.73
MM1 pKa = 7.89PDD3 pKa = 3.61DD4 pKa = 4.49VDD6 pKa = 4.33MKK8 pKa = 9.84STCNVDD14 pKa = 3.13GNQVTKK20 pKa = 11.32NKK22 pKa = 8.86MFSGNWMVVMEE33 pKa = 4.76GLTFAWMRR41 pKa = 11.84TFEE44 pKa = 4.38IVAGPPVTITSTPTATYY61 pKa = 8.99TITSTPRR68 pKa = 11.84ATVTTTSTDD77 pKa = 2.77IYY79 pKa = 10.13STTLSPLTVTVPSSTSTRR97 pKa = 11.84TITTTPKK104 pKa = 10.27QVTVDD109 pKa = 3.54STSTSTRR116 pKa = 11.84TRR118 pKa = 11.84ISRR121 pKa = 11.84TWSATVTTTTVTVSCQPQIVKK142 pKa = 10.09RR143 pKa = 11.84DD144 pKa = 3.75PTCTIRR150 pKa = 11.84PTKK153 pKa = 9.61ATLAAASNTTYY164 pKa = 9.78PAPRR168 pKa = 11.84GRR170 pKa = 11.84FFRR173 pKa = 11.84GAPRR177 pKa = 11.84RR178 pKa = 11.84SRR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84EE183 pKa = 3.72AFVEE187 pKa = 3.94PRR189 pKa = 11.84RR190 pKa = 11.84EE191 pKa = 3.83NQLFEE196 pKa = 5.0RR197 pKa = 11.84GAGTLLL203 pKa = 3.73
Molecular weight: 22.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5944843 |
49 |
5561 |
493.5 |
54.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.605 ± 0.016 | 1.219 ± 0.007 |
5.773 ± 0.014 | 6.004 ± 0.019 |
3.755 ± 0.012 | 6.868 ± 0.022 |
2.461 ± 0.011 | 4.836 ± 0.014 |
4.675 ± 0.018 | 8.989 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.158 ± 0.008 | 3.566 ± 0.01 |
5.924 ± 0.022 | 4.061 ± 0.015 |
6.165 ± 0.018 | 8.31 ± 0.023 |
6.084 ± 0.016 | 6.273 ± 0.016 |
1.491 ± 0.008 | 2.782 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
