
Clostridiales bacterium
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; unclassified Eubacteriales
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
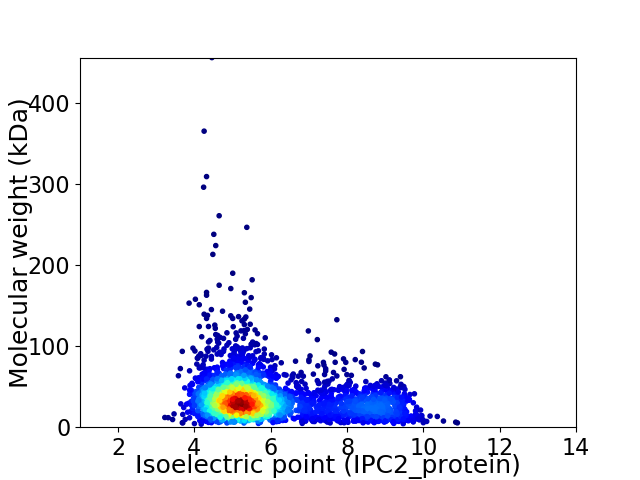
Virtual 2D-PAGE plot for 3011 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
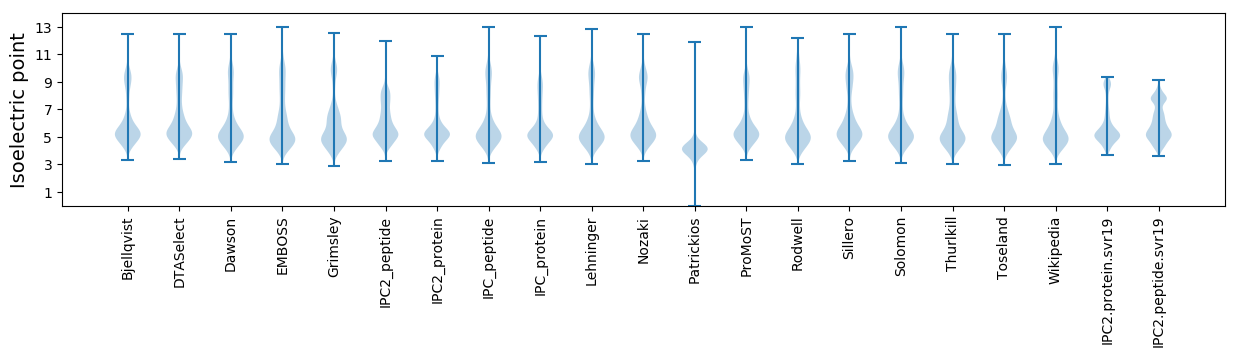
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W2AYN2|A0A1W2AYN2_9FIRM Diguanylate cyclase (GGDEF) domain-containing protein OS=Clostridiales bacterium OX=1898207 GN=SAMN06297397_1809 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.62KK3 pKa = 8.91MLCFILAAAMMFSITALAEE22 pKa = 4.34DD23 pKa = 4.73SPGGTPPDD31 pKa = 4.2GFSGTPPEE39 pKa = 4.47GTPPDD44 pKa = 3.87GFGGGTPPDD53 pKa = 3.83GTPPDD58 pKa = 3.94GFGGGTPPDD67 pKa = 4.06GAPGGFGGGTPPGGGTSSFEE87 pKa = 4.05YY88 pKa = 10.47AASTEE93 pKa = 4.2VTEE96 pKa = 4.27AAEE99 pKa = 4.36LNSQSFVSEE108 pKa = 4.19TADD111 pKa = 3.79EE112 pKa = 4.4SALLINTADD121 pKa = 3.32TVTITEE127 pKa = 4.03PAITKK132 pKa = 9.46TGDD135 pKa = 2.94SSGGDD140 pKa = 2.97NCNFYY145 pKa = 11.2GLNAALLVMGGSTTTITGGTIEE167 pKa = 4.47TSASGANGVFSYY179 pKa = 10.76GGNGGRR185 pKa = 11.84NGAAGDD191 pKa = 3.74GTTVVISDD199 pKa = 3.59TTIVTDD205 pKa = 3.83GDD207 pKa = 3.93GSGGIMTTGGGITYY221 pKa = 10.38ADD223 pKa = 3.75NLAVTTSGRR232 pKa = 11.84SSAAIRR238 pKa = 11.84TDD240 pKa = 3.07RR241 pKa = 11.84GGGTVVVDD249 pKa = 3.78GGTYY253 pKa = 6.18TTSGLGSPAIYY264 pKa = 8.92STADD268 pKa = 2.89ITVNNASLVSNLSEE282 pKa = 4.28GVCIEE287 pKa = 3.9GMNSITLEE295 pKa = 3.99NSNLTASNTQCNGNATFHH313 pKa = 6.17DD314 pKa = 4.5TIMIYY319 pKa = 10.77QSMSGDD325 pKa = 3.59ADD327 pKa = 3.69SGTSSFTMTGGSLTSLSGHH346 pKa = 5.84MFHH349 pKa = 6.55VTNTHH354 pKa = 6.77AVITLNGVEE363 pKa = 4.96LSNQGSDD370 pKa = 2.98ILLSVCDD377 pKa = 4.98DD378 pKa = 3.55GWHH381 pKa = 6.08GASNVAEE388 pKa = 4.2LVTHH392 pKa = 5.94NQQLEE397 pKa = 4.26GTILAGDD404 pKa = 3.69NSVLKK409 pKa = 9.56LTLSDD414 pKa = 3.64GSAFEE419 pKa = 4.38GTVSGRR425 pKa = 11.84IEE427 pKa = 3.81NAKK430 pKa = 10.52NEE432 pKa = 4.49LISTEE437 pKa = 4.08VGSVSVTLDD446 pKa = 3.33STSTWTLTADD456 pKa = 3.12TWIDD460 pKa = 3.4AFEE463 pKa = 5.05GDD465 pKa = 3.55ASRR468 pKa = 11.84VTGNGYY474 pKa = 7.02TLYY477 pKa = 11.31VNGTALEE484 pKa = 4.97GISS487 pKa = 3.56
MM1 pKa = 7.62KK2 pKa = 10.62KK3 pKa = 8.91MLCFILAAAMMFSITALAEE22 pKa = 4.34DD23 pKa = 4.73SPGGTPPDD31 pKa = 4.2GFSGTPPEE39 pKa = 4.47GTPPDD44 pKa = 3.87GFGGGTPPDD53 pKa = 3.83GTPPDD58 pKa = 3.94GFGGGTPPDD67 pKa = 4.06GAPGGFGGGTPPGGGTSSFEE87 pKa = 4.05YY88 pKa = 10.47AASTEE93 pKa = 4.2VTEE96 pKa = 4.27AAEE99 pKa = 4.36LNSQSFVSEE108 pKa = 4.19TADD111 pKa = 3.79EE112 pKa = 4.4SALLINTADD121 pKa = 3.32TVTITEE127 pKa = 4.03PAITKK132 pKa = 9.46TGDD135 pKa = 2.94SSGGDD140 pKa = 2.97NCNFYY145 pKa = 11.2GLNAALLVMGGSTTTITGGTIEE167 pKa = 4.47TSASGANGVFSYY179 pKa = 10.76GGNGGRR185 pKa = 11.84NGAAGDD191 pKa = 3.74GTTVVISDD199 pKa = 3.59TTIVTDD205 pKa = 3.83GDD207 pKa = 3.93GSGGIMTTGGGITYY221 pKa = 10.38ADD223 pKa = 3.75NLAVTTSGRR232 pKa = 11.84SSAAIRR238 pKa = 11.84TDD240 pKa = 3.07RR241 pKa = 11.84GGGTVVVDD249 pKa = 3.78GGTYY253 pKa = 6.18TTSGLGSPAIYY264 pKa = 8.92STADD268 pKa = 2.89ITVNNASLVSNLSEE282 pKa = 4.28GVCIEE287 pKa = 3.9GMNSITLEE295 pKa = 3.99NSNLTASNTQCNGNATFHH313 pKa = 6.17DD314 pKa = 4.5TIMIYY319 pKa = 10.77QSMSGDD325 pKa = 3.59ADD327 pKa = 3.69SGTSSFTMTGGSLTSLSGHH346 pKa = 5.84MFHH349 pKa = 6.55VTNTHH354 pKa = 6.77AVITLNGVEE363 pKa = 4.96LSNQGSDD370 pKa = 2.98ILLSVCDD377 pKa = 4.98DD378 pKa = 3.55GWHH381 pKa = 6.08GASNVAEE388 pKa = 4.2LVTHH392 pKa = 5.94NQQLEE397 pKa = 4.26GTILAGDD404 pKa = 3.69NSVLKK409 pKa = 9.56LTLSDD414 pKa = 3.64GSAFEE419 pKa = 4.38GTVSGRR425 pKa = 11.84IEE427 pKa = 3.81NAKK430 pKa = 10.52NEE432 pKa = 4.49LISTEE437 pKa = 4.08VGSVSVTLDD446 pKa = 3.33STSTWTLTADD456 pKa = 3.12TWIDD460 pKa = 3.4AFEE463 pKa = 5.05GDD465 pKa = 3.55ASRR468 pKa = 11.84VTGNGYY474 pKa = 7.02TLYY477 pKa = 11.31VNGTALEE484 pKa = 4.97GISS487 pKa = 3.56
Molecular weight: 48.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W2BXG4|A0A1W2BXG4_9FIRM Phosphate propanoyltransferase OS=Clostridiales bacterium OX=1898207 GN=SAMN06297397_2445 PE=3 SV=1
MM1 pKa = 7.25FRR3 pKa = 11.84TYY5 pKa = 9.97QPKK8 pKa = 9.54KK9 pKa = 7.56RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.87GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.97NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.64EE41 pKa = 3.53LTVV44 pKa = 3.27
MM1 pKa = 7.25FRR3 pKa = 11.84TYY5 pKa = 9.97QPKK8 pKa = 9.54KK9 pKa = 7.56RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.87GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.97NGRR28 pKa = 11.84RR29 pKa = 11.84VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.64EE41 pKa = 3.53LTVV44 pKa = 3.27
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1014911 |
34 |
4155 |
337.1 |
37.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.436 ± 0.053 | 1.549 ± 0.019 |
5.928 ± 0.034 | 7.477 ± 0.057 |
3.895 ± 0.03 | 7.459 ± 0.043 |
1.749 ± 0.018 | 6.307 ± 0.045 |
5.414 ± 0.046 | 9.327 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.019 ± 0.025 | 3.877 ± 0.034 |
4.07 ± 0.028 | 2.977 ± 0.023 |
5.33 ± 0.041 | 5.552 ± 0.038 |
5.825 ± 0.049 | 6.77 ± 0.033 |
1.231 ± 0.017 | 3.808 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
