
Culex rhabdo-like virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ohlsrhavirus; Culex ohlsrhavirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2RTB8|A0A1Z2RTB8_9RHAB Glycoprotein OS=Culex rhabdo-like virus OX=2010277 PE=4 SV=1
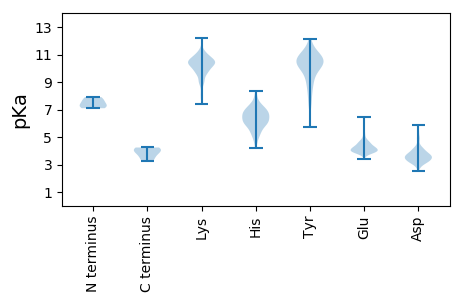
MM1 pKa = 7.2IHH3 pKa = 6.8NSEE6 pKa = 4.25LLANAFLMIALISWTTCEE24 pKa = 4.16DD25 pKa = 3.01QSHH28 pKa = 5.33NMKK31 pKa = 10.21ILFPTSSKK39 pKa = 10.69IEE41 pKa = 3.84WHH43 pKa = 6.77DD44 pKa = 3.61VSPSNLICSTARR56 pKa = 11.84IHH58 pKa = 6.72QNIAHH63 pKa = 6.64GLSFVVNRR71 pKa = 11.84PVIGKK76 pKa = 9.18DD77 pKa = 3.16PKK79 pKa = 10.65ISGTLCVAIEE89 pKa = 4.03LTTTCSKK96 pKa = 11.22GFFGGVDD103 pKa = 3.6VTLSTSPADD112 pKa = 3.93LSEE115 pKa = 4.17EE116 pKa = 4.04NCRR119 pKa = 11.84KK120 pKa = 10.12EE121 pKa = 4.47IASQNQGEE129 pKa = 4.54FSSAEE134 pKa = 3.97HH135 pKa = 6.02PQPVCSWMKK144 pKa = 9.63TSSTSRR150 pKa = 11.84TVIHH154 pKa = 6.73LSKK157 pKa = 10.1MDD159 pKa = 3.47AHH161 pKa = 6.94YY162 pKa = 11.0DD163 pKa = 3.73PYY165 pKa = 11.47SDD167 pKa = 4.16TLFSSIFLSGKK178 pKa = 9.55CRR180 pKa = 11.84ASFCQTVFEE189 pKa = 4.14NRR191 pKa = 11.84LWIPQEE197 pKa = 4.11SLTEE201 pKa = 4.12YY202 pKa = 10.62CSTEE206 pKa = 3.88HH207 pKa = 6.71LVEE210 pKa = 4.32SSMMIYY216 pKa = 10.38NSSSNGSSLWSPDD229 pKa = 2.86FDD231 pKa = 3.36ISEE234 pKa = 4.24EE235 pKa = 4.01EE236 pKa = 4.72KK237 pKa = 9.93PCIMNFCGQKK247 pKa = 10.27GLRR250 pKa = 11.84FASGEE255 pKa = 3.98WVALSRR261 pKa = 11.84QSIPKK266 pKa = 9.18APWVGEE272 pKa = 4.09HH273 pKa = 6.92FYY275 pKa = 11.0HH276 pKa = 7.51LKK278 pKa = 10.87DD279 pKa = 3.91CDD281 pKa = 3.28GHH283 pKa = 4.85TTVNLVEE290 pKa = 3.91AHH292 pKa = 5.8QDD294 pKa = 3.23LRR296 pKa = 11.84HH297 pKa = 4.75MTKK300 pKa = 10.01VVLEE304 pKa = 4.11EE305 pKa = 5.56FIDD308 pKa = 4.17EE309 pKa = 4.2QCEE312 pKa = 4.01MTVNKK317 pKa = 10.21LKK319 pKa = 10.04EE320 pKa = 3.98GNMISRR326 pKa = 11.84IEE328 pKa = 4.07LQTLYY333 pKa = 10.36PRR335 pKa = 11.84TPGFHH340 pKa = 7.03PIYY343 pKa = 10.02RR344 pKa = 11.84YY345 pKa = 7.48TQGKK349 pKa = 8.81FLMGMAHH356 pKa = 6.02YY357 pKa = 9.48SWVSIEE363 pKa = 3.96SSEE366 pKa = 3.85IFPYY370 pKa = 10.37ISIRR374 pKa = 11.84SSLNKK379 pKa = 10.21SIEE382 pKa = 3.96YY383 pKa = 10.46SYY385 pKa = 8.35WTMDD389 pKa = 2.86NRR391 pKa = 11.84TQIIDD396 pKa = 4.1GPNGLYY402 pKa = 10.16ILDD405 pKa = 3.83HH406 pKa = 5.28QLIYY410 pKa = 10.87GLEE413 pKa = 4.03EE414 pKa = 3.64EE415 pKa = 4.46QKK417 pKa = 10.43FRR419 pKa = 11.84RR420 pKa = 11.84ILSKK424 pKa = 10.43SSKK427 pKa = 10.12HH428 pKa = 5.23VFPILPEE435 pKa = 3.91QKK437 pKa = 9.37TNGQSFVRR445 pKa = 11.84LLSHH449 pKa = 6.53TSYY452 pKa = 11.2SFSDD456 pKa = 3.8DD457 pKa = 3.28ASLVDD462 pKa = 4.92VIWHH466 pKa = 6.32PVLTCLIITAILLPSMIGIIWCTKK490 pKa = 8.19NQVFIKK496 pKa = 9.94KK497 pKa = 9.61LKK499 pKa = 10.57SFAQRR504 pKa = 11.84DD505 pKa = 3.89VEE507 pKa = 4.0PSEE510 pKa = 5.42DD511 pKa = 3.21ISEE514 pKa = 4.54AEE516 pKa = 3.88GEE518 pKa = 4.47YY519 pKa = 10.78FNPP522 pKa = 4.24
MM1 pKa = 7.2IHH3 pKa = 6.8NSEE6 pKa = 4.25LLANAFLMIALISWTTCEE24 pKa = 4.16DD25 pKa = 3.01QSHH28 pKa = 5.33NMKK31 pKa = 10.21ILFPTSSKK39 pKa = 10.69IEE41 pKa = 3.84WHH43 pKa = 6.77DD44 pKa = 3.61VSPSNLICSTARR56 pKa = 11.84IHH58 pKa = 6.72QNIAHH63 pKa = 6.64GLSFVVNRR71 pKa = 11.84PVIGKK76 pKa = 9.18DD77 pKa = 3.16PKK79 pKa = 10.65ISGTLCVAIEE89 pKa = 4.03LTTTCSKK96 pKa = 11.22GFFGGVDD103 pKa = 3.6VTLSTSPADD112 pKa = 3.93LSEE115 pKa = 4.17EE116 pKa = 4.04NCRR119 pKa = 11.84KK120 pKa = 10.12EE121 pKa = 4.47IASQNQGEE129 pKa = 4.54FSSAEE134 pKa = 3.97HH135 pKa = 6.02PQPVCSWMKK144 pKa = 9.63TSSTSRR150 pKa = 11.84TVIHH154 pKa = 6.73LSKK157 pKa = 10.1MDD159 pKa = 3.47AHH161 pKa = 6.94YY162 pKa = 11.0DD163 pKa = 3.73PYY165 pKa = 11.47SDD167 pKa = 4.16TLFSSIFLSGKK178 pKa = 9.55CRR180 pKa = 11.84ASFCQTVFEE189 pKa = 4.14NRR191 pKa = 11.84LWIPQEE197 pKa = 4.11SLTEE201 pKa = 4.12YY202 pKa = 10.62CSTEE206 pKa = 3.88HH207 pKa = 6.71LVEE210 pKa = 4.32SSMMIYY216 pKa = 10.38NSSSNGSSLWSPDD229 pKa = 2.86FDD231 pKa = 3.36ISEE234 pKa = 4.24EE235 pKa = 4.01EE236 pKa = 4.72KK237 pKa = 9.93PCIMNFCGQKK247 pKa = 10.27GLRR250 pKa = 11.84FASGEE255 pKa = 3.98WVALSRR261 pKa = 11.84QSIPKK266 pKa = 9.18APWVGEE272 pKa = 4.09HH273 pKa = 6.92FYY275 pKa = 11.0HH276 pKa = 7.51LKK278 pKa = 10.87DD279 pKa = 3.91CDD281 pKa = 3.28GHH283 pKa = 4.85TTVNLVEE290 pKa = 3.91AHH292 pKa = 5.8QDD294 pKa = 3.23LRR296 pKa = 11.84HH297 pKa = 4.75MTKK300 pKa = 10.01VVLEE304 pKa = 4.11EE305 pKa = 5.56FIDD308 pKa = 4.17EE309 pKa = 4.2QCEE312 pKa = 4.01MTVNKK317 pKa = 10.21LKK319 pKa = 10.04EE320 pKa = 3.98GNMISRR326 pKa = 11.84IEE328 pKa = 4.07LQTLYY333 pKa = 10.36PRR335 pKa = 11.84TPGFHH340 pKa = 7.03PIYY343 pKa = 10.02RR344 pKa = 11.84YY345 pKa = 7.48TQGKK349 pKa = 8.81FLMGMAHH356 pKa = 6.02YY357 pKa = 9.48SWVSIEE363 pKa = 3.96SSEE366 pKa = 3.85IFPYY370 pKa = 10.37ISIRR374 pKa = 11.84SSLNKK379 pKa = 10.21SIEE382 pKa = 3.96YY383 pKa = 10.46SYY385 pKa = 8.35WTMDD389 pKa = 2.86NRR391 pKa = 11.84TQIIDD396 pKa = 4.1GPNGLYY402 pKa = 10.16ILDD405 pKa = 3.83HH406 pKa = 5.28QLIYY410 pKa = 10.87GLEE413 pKa = 4.03EE414 pKa = 3.64EE415 pKa = 4.46QKK417 pKa = 10.43FRR419 pKa = 11.84RR420 pKa = 11.84ILSKK424 pKa = 10.43SSKK427 pKa = 10.12HH428 pKa = 5.23VFPILPEE435 pKa = 3.91QKK437 pKa = 9.37TNGQSFVRR445 pKa = 11.84LLSHH449 pKa = 6.53TSYY452 pKa = 11.2SFSDD456 pKa = 3.8DD457 pKa = 3.28ASLVDD462 pKa = 4.92VIWHH466 pKa = 6.32PVLTCLIITAILLPSMIGIIWCTKK490 pKa = 8.19NQVFIKK496 pKa = 9.94KK497 pKa = 9.61LKK499 pKa = 10.57SFAQRR504 pKa = 11.84DD505 pKa = 3.89VEE507 pKa = 4.0PSEE510 pKa = 5.42DD511 pKa = 3.21ISEE514 pKa = 4.54AEE516 pKa = 3.88GEE518 pKa = 4.47YY519 pKa = 10.78FNPP522 pKa = 4.24
Molecular weight: 59.29 kDa
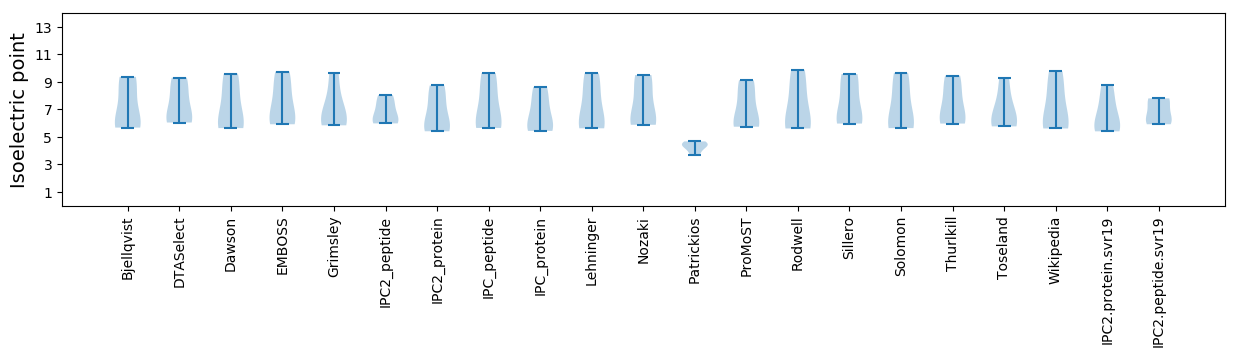
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RT82|A0A1Z2RT82_9RHAB Nucleocapsid protein OS=Culex rhabdo-like virus OX=2010277 PE=4 SV=1
MM1 pKa = 7.39NNSTQIKK8 pKa = 9.75KK9 pKa = 8.53KK10 pKa = 8.03TFNMCVSKK18 pKa = 11.04VLLVNATIEE27 pKa = 3.98IKK29 pKa = 10.6YY30 pKa = 10.13RR31 pKa = 11.84NPPKK35 pKa = 9.49TCKK38 pKa = 10.07EE39 pKa = 4.32LIHH42 pKa = 7.48SLEE45 pKa = 4.52LIKK48 pKa = 11.12DD49 pKa = 3.53GGNWYY54 pKa = 10.43QSDD57 pKa = 3.68QEE59 pKa = 4.69FIFFFYY65 pKa = 10.42ILSGYY70 pKa = 6.08TCKK73 pKa = 10.21HH74 pKa = 4.2VHH76 pKa = 6.1HH77 pKa = 7.16LGGFHH82 pKa = 7.89LYY84 pKa = 10.0RR85 pKa = 11.84GCISGAFEE93 pKa = 4.34IKK95 pKa = 10.35FKK97 pKa = 10.85GISPRR102 pKa = 11.84FPLTYY107 pKa = 9.44EE108 pKa = 4.0YY109 pKa = 10.49QGEE112 pKa = 4.5GLGMIPDD119 pKa = 4.08TLISFSLVAEE129 pKa = 4.41EE130 pKa = 4.83SLVSPISLWGYY141 pKa = 9.04IIKK144 pKa = 10.61GSPSKK149 pKa = 10.5YY150 pKa = 9.25RR151 pKa = 11.84KK152 pKa = 9.81SNEE155 pKa = 3.93IKK157 pKa = 10.05WGIFDD162 pKa = 4.55NFPLSVKK169 pKa = 10.41GKK171 pKa = 10.35DD172 pKa = 3.41GIFTIRR178 pKa = 11.84RR179 pKa = 11.84KK180 pKa = 7.73TT181 pKa = 3.26
MM1 pKa = 7.39NNSTQIKK8 pKa = 9.75KK9 pKa = 8.53KK10 pKa = 8.03TFNMCVSKK18 pKa = 11.04VLLVNATIEE27 pKa = 3.98IKK29 pKa = 10.6YY30 pKa = 10.13RR31 pKa = 11.84NPPKK35 pKa = 9.49TCKK38 pKa = 10.07EE39 pKa = 4.32LIHH42 pKa = 7.48SLEE45 pKa = 4.52LIKK48 pKa = 11.12DD49 pKa = 3.53GGNWYY54 pKa = 10.43QSDD57 pKa = 3.68QEE59 pKa = 4.69FIFFFYY65 pKa = 10.42ILSGYY70 pKa = 6.08TCKK73 pKa = 10.21HH74 pKa = 4.2VHH76 pKa = 6.1HH77 pKa = 7.16LGGFHH82 pKa = 7.89LYY84 pKa = 10.0RR85 pKa = 11.84GCISGAFEE93 pKa = 4.34IKK95 pKa = 10.35FKK97 pKa = 10.85GISPRR102 pKa = 11.84FPLTYY107 pKa = 9.44EE108 pKa = 4.0YY109 pKa = 10.49QGEE112 pKa = 4.5GLGMIPDD119 pKa = 4.08TLISFSLVAEE129 pKa = 4.41EE130 pKa = 4.83SLVSPISLWGYY141 pKa = 9.04IIKK144 pKa = 10.61GSPSKK149 pKa = 10.5YY150 pKa = 9.25RR151 pKa = 11.84KK152 pKa = 9.81SNEE155 pKa = 3.93IKK157 pKa = 10.05WGIFDD162 pKa = 4.55NFPLSVKK169 pKa = 10.41GKK171 pKa = 10.35DD172 pKa = 3.41GIFTIRR178 pKa = 11.84RR179 pKa = 11.84KK180 pKa = 7.73TT181 pKa = 3.26
Molecular weight: 20.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3665 |
181 |
2119 |
733.0 |
83.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.73 ± 0.798 | 1.583 ± 0.348 |
5.293 ± 0.533 | 6.276 ± 0.516 |
4.475 ± 0.41 | 5.812 ± 0.458 |
2.456 ± 0.26 | 6.603 ± 0.633 |
6.439 ± 0.749 | 9.168 ± 0.811 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.647 ± 0.221 | 4.529 ± 0.29 |
4.748 ± 0.465 | 3.356 ± 0.294 |
5.184 ± 0.609 | 8.759 ± 0.733 |
5.703 ± 0.274 | 5.921 ± 0.34 |
1.692 ± 0.201 | 3.629 ± 0.314 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
