
Mengla dianlovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Filoviridae; Dianlovirus
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins
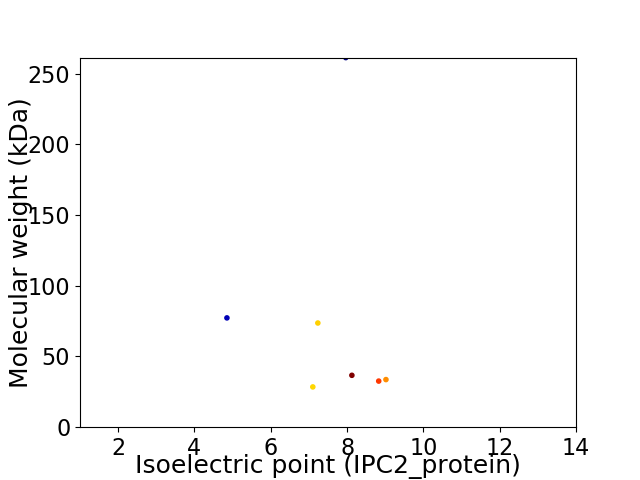
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U6Q5|A0A3Q8U6Q5_9MONO Matrix protein VP40 OS=Mengla dianlovirus OX=2496529 GN=VP40 PE=3 SV=1
MM1 pKa = 8.02DD2 pKa = 4.37LHH4 pKa = 6.96GLLEE8 pKa = 4.89LGTRR12 pKa = 11.84PTAPHH17 pKa = 4.84VRR19 pKa = 11.84SRR21 pKa = 11.84KK22 pKa = 8.5IVIYY26 pKa = 8.3EE27 pKa = 3.97TGNQIIICNQIIDD40 pKa = 4.07AVSAGIDD47 pKa = 3.76LGDD50 pKa = 3.98LLEE53 pKa = 4.96GCLLTLCLEE62 pKa = 4.65HH63 pKa = 7.61YY64 pKa = 10.19YY65 pKa = 11.2GSDD68 pKa = 3.04KK69 pKa = 11.6DD70 pKa = 4.13KK71 pKa = 11.3FNSSQMAAYY80 pKa = 10.18LRR82 pKa = 11.84DD83 pKa = 3.25AGYY86 pKa = 10.78DD87 pKa = 3.44FEE89 pKa = 5.48VIRR92 pKa = 11.84AQDD95 pKa = 3.4AKK97 pKa = 10.87KK98 pKa = 10.39LADD101 pKa = 5.04LIPRR105 pKa = 11.84EE106 pKa = 4.14SHH108 pKa = 6.13LLNVISALEE117 pKa = 3.95NLDD120 pKa = 3.5GSEE123 pKa = 4.15KK124 pKa = 10.26NKK126 pKa = 9.95QRR128 pKa = 11.84VGLFLSFCSLFLPKK142 pKa = 10.43LVVGDD147 pKa = 3.63KK148 pKa = 11.22ASIEE152 pKa = 3.94KK153 pKa = 10.52ALRR156 pKa = 11.84QVAIHH161 pKa = 5.78QEE163 pKa = 3.96QGMVVYY169 pKa = 10.01PPTWLTTGFMKK180 pKa = 11.06LIFSIVRR187 pKa = 11.84ASFIVKK193 pKa = 9.59FVLIHH198 pKa = 5.68QGINLVTGHH207 pKa = 7.28DD208 pKa = 3.75AYY210 pKa = 11.29DD211 pKa = 3.64NVISNSINQTRR222 pKa = 11.84FSGLLIVKK230 pKa = 8.18TVLEE234 pKa = 5.12HH235 pKa = 6.31ILQRR239 pKa = 11.84TEE241 pKa = 3.61NGVILHH247 pKa = 6.07PLVRR251 pKa = 11.84TSKK254 pKa = 10.25VKK256 pKa = 10.96SEE258 pKa = 4.06VEE260 pKa = 4.11SFKK263 pKa = 11.08VALRR267 pKa = 11.84GLARR271 pKa = 11.84HH272 pKa = 6.19KK273 pKa = 10.44EE274 pKa = 4.0YY275 pKa = 11.22APFARR280 pKa = 11.84VLNLSGVNNLEE291 pKa = 4.48HH292 pKa = 7.06GLFPQLSAIAIGVATAHH309 pKa = 6.61GSTLAGVSVGEE320 pKa = 4.2QYY322 pKa = 10.54QQLRR326 pKa = 11.84EE327 pKa = 4.08AAHH330 pKa = 6.28DD331 pKa = 4.19AEE333 pKa = 5.43LKK335 pKa = 9.0LQRR338 pKa = 11.84RR339 pKa = 11.84RR340 pKa = 11.84EE341 pKa = 3.91QMEE344 pKa = 3.96ISSLEE349 pKa = 3.97LDD351 pKa = 4.17LEE353 pKa = 4.28EE354 pKa = 5.19QKK356 pKa = 10.75ILEE359 pKa = 4.18QFHH362 pKa = 4.9QQKK365 pKa = 8.7TEE367 pKa = 3.68ITHH370 pKa = 5.13TQTLAVLTQKK380 pKa = 10.61KK381 pKa = 9.65EE382 pKa = 4.07KK383 pKa = 9.72LAKK386 pKa = 10.29LAGEE390 pKa = 4.58LGADD394 pKa = 3.52KK395 pKa = 10.74LFQPVIPQPNDD406 pKa = 3.02KK407 pKa = 10.33PQMTTVIDD415 pKa = 3.95PSRR418 pKa = 11.84TSVKK422 pKa = 9.82IQTSFLPPPSTKK434 pKa = 10.96DD435 pKa = 3.24NMEE438 pKa = 3.92GQIPEE443 pKa = 4.35SPEE446 pKa = 3.64SSTISSSSSCIDD458 pKa = 3.45PNDD461 pKa = 3.82PFALLIDD468 pKa = 4.79DD469 pKa = 5.5DD470 pKa = 4.22EE471 pKa = 5.13QEE473 pKa = 4.99GDD475 pKa = 3.76FQSADD480 pKa = 3.49EE481 pKa = 5.35GDD483 pKa = 4.22SQDD486 pKa = 3.66QSAEE490 pKa = 3.66AARR493 pKa = 11.84QEE495 pKa = 4.5EE496 pKa = 4.35IRR498 pKa = 11.84KK499 pKa = 9.83AEE501 pKa = 4.1QKK503 pKa = 10.77FKK505 pKa = 10.85GLPRR509 pKa = 11.84TIPTKK514 pKa = 10.51DD515 pKa = 3.21QNPPIKK521 pKa = 9.89PQRR524 pKa = 11.84TNLAPVQEE532 pKa = 4.52EE533 pKa = 4.58SEE535 pKa = 4.43SEE537 pKa = 4.17YY538 pKa = 8.68TTTSDD543 pKa = 3.12EE544 pKa = 4.43SGSEE548 pKa = 4.32DD549 pKa = 3.58EE550 pKa = 6.64SSDD553 pKa = 3.27QRR555 pKa = 11.84NIEE558 pKa = 4.21VPPPPLYY565 pKa = 9.69DD566 pKa = 3.26TSTSEE571 pKa = 4.06IGDD574 pKa = 3.43MGAAEE579 pKa = 4.29HH580 pKa = 6.2TLDD583 pKa = 4.65PFGDD587 pKa = 4.02MPPLEE592 pKa = 4.59TDD594 pKa = 3.27VLTPQDD600 pKa = 3.59TATVPSAPPSSPTPSTNRR618 pKa = 11.84QGEE621 pKa = 4.08AQTQNGEE628 pKa = 4.54DD629 pKa = 4.33SSQDD633 pKa = 2.43WPRR636 pKa = 11.84RR637 pKa = 11.84VKK639 pKa = 9.54TNKK642 pKa = 9.17GRR644 pKa = 11.84EE645 pKa = 4.03FMFPTDD651 pKa = 3.98LLHH654 pKa = 6.23RR655 pKa = 11.84TPPQVLLDD663 pKa = 3.79ALVNEE668 pKa = 4.66YY669 pKa = 10.29EE670 pKa = 4.56SPLSATEE677 pKa = 5.73LSDD680 pKa = 4.09DD681 pKa = 3.6WPEE684 pKa = 3.75MTFEE688 pKa = 3.98EE689 pKa = 4.57RR690 pKa = 11.84KK691 pKa = 10.11NVAFNLL697 pKa = 3.9
MM1 pKa = 8.02DD2 pKa = 4.37LHH4 pKa = 6.96GLLEE8 pKa = 4.89LGTRR12 pKa = 11.84PTAPHH17 pKa = 4.84VRR19 pKa = 11.84SRR21 pKa = 11.84KK22 pKa = 8.5IVIYY26 pKa = 8.3EE27 pKa = 3.97TGNQIIICNQIIDD40 pKa = 4.07AVSAGIDD47 pKa = 3.76LGDD50 pKa = 3.98LLEE53 pKa = 4.96GCLLTLCLEE62 pKa = 4.65HH63 pKa = 7.61YY64 pKa = 10.19YY65 pKa = 11.2GSDD68 pKa = 3.04KK69 pKa = 11.6DD70 pKa = 4.13KK71 pKa = 11.3FNSSQMAAYY80 pKa = 10.18LRR82 pKa = 11.84DD83 pKa = 3.25AGYY86 pKa = 10.78DD87 pKa = 3.44FEE89 pKa = 5.48VIRR92 pKa = 11.84AQDD95 pKa = 3.4AKK97 pKa = 10.87KK98 pKa = 10.39LADD101 pKa = 5.04LIPRR105 pKa = 11.84EE106 pKa = 4.14SHH108 pKa = 6.13LLNVISALEE117 pKa = 3.95NLDD120 pKa = 3.5GSEE123 pKa = 4.15KK124 pKa = 10.26NKK126 pKa = 9.95QRR128 pKa = 11.84VGLFLSFCSLFLPKK142 pKa = 10.43LVVGDD147 pKa = 3.63KK148 pKa = 11.22ASIEE152 pKa = 3.94KK153 pKa = 10.52ALRR156 pKa = 11.84QVAIHH161 pKa = 5.78QEE163 pKa = 3.96QGMVVYY169 pKa = 10.01PPTWLTTGFMKK180 pKa = 11.06LIFSIVRR187 pKa = 11.84ASFIVKK193 pKa = 9.59FVLIHH198 pKa = 5.68QGINLVTGHH207 pKa = 7.28DD208 pKa = 3.75AYY210 pKa = 11.29DD211 pKa = 3.64NVISNSINQTRR222 pKa = 11.84FSGLLIVKK230 pKa = 8.18TVLEE234 pKa = 5.12HH235 pKa = 6.31ILQRR239 pKa = 11.84TEE241 pKa = 3.61NGVILHH247 pKa = 6.07PLVRR251 pKa = 11.84TSKK254 pKa = 10.25VKK256 pKa = 10.96SEE258 pKa = 4.06VEE260 pKa = 4.11SFKK263 pKa = 11.08VALRR267 pKa = 11.84GLARR271 pKa = 11.84HH272 pKa = 6.19KK273 pKa = 10.44EE274 pKa = 4.0YY275 pKa = 11.22APFARR280 pKa = 11.84VLNLSGVNNLEE291 pKa = 4.48HH292 pKa = 7.06GLFPQLSAIAIGVATAHH309 pKa = 6.61GSTLAGVSVGEE320 pKa = 4.2QYY322 pKa = 10.54QQLRR326 pKa = 11.84EE327 pKa = 4.08AAHH330 pKa = 6.28DD331 pKa = 4.19AEE333 pKa = 5.43LKK335 pKa = 9.0LQRR338 pKa = 11.84RR339 pKa = 11.84RR340 pKa = 11.84EE341 pKa = 3.91QMEE344 pKa = 3.96ISSLEE349 pKa = 3.97LDD351 pKa = 4.17LEE353 pKa = 4.28EE354 pKa = 5.19QKK356 pKa = 10.75ILEE359 pKa = 4.18QFHH362 pKa = 4.9QQKK365 pKa = 8.7TEE367 pKa = 3.68ITHH370 pKa = 5.13TQTLAVLTQKK380 pKa = 10.61KK381 pKa = 9.65EE382 pKa = 4.07KK383 pKa = 9.72LAKK386 pKa = 10.29LAGEE390 pKa = 4.58LGADD394 pKa = 3.52KK395 pKa = 10.74LFQPVIPQPNDD406 pKa = 3.02KK407 pKa = 10.33PQMTTVIDD415 pKa = 3.95PSRR418 pKa = 11.84TSVKK422 pKa = 9.82IQTSFLPPPSTKK434 pKa = 10.96DD435 pKa = 3.24NMEE438 pKa = 3.92GQIPEE443 pKa = 4.35SPEE446 pKa = 3.64SSTISSSSSCIDD458 pKa = 3.45PNDD461 pKa = 3.82PFALLIDD468 pKa = 4.79DD469 pKa = 5.5DD470 pKa = 4.22EE471 pKa = 5.13QEE473 pKa = 4.99GDD475 pKa = 3.76FQSADD480 pKa = 3.49EE481 pKa = 5.35GDD483 pKa = 4.22SQDD486 pKa = 3.66QSAEE490 pKa = 3.66AARR493 pKa = 11.84QEE495 pKa = 4.5EE496 pKa = 4.35IRR498 pKa = 11.84KK499 pKa = 9.83AEE501 pKa = 4.1QKK503 pKa = 10.77FKK505 pKa = 10.85GLPRR509 pKa = 11.84TIPTKK514 pKa = 10.51DD515 pKa = 3.21QNPPIKK521 pKa = 9.89PQRR524 pKa = 11.84TNLAPVQEE532 pKa = 4.52EE533 pKa = 4.58SEE535 pKa = 4.43SEE537 pKa = 4.17YY538 pKa = 8.68TTTSDD543 pKa = 3.12EE544 pKa = 4.43SGSEE548 pKa = 4.32DD549 pKa = 3.58EE550 pKa = 6.64SSDD553 pKa = 3.27QRR555 pKa = 11.84NIEE558 pKa = 4.21VPPPPLYY565 pKa = 9.69DD566 pKa = 3.26TSTSEE571 pKa = 4.06IGDD574 pKa = 3.43MGAAEE579 pKa = 4.29HH580 pKa = 6.2TLDD583 pKa = 4.65PFGDD587 pKa = 4.02MPPLEE592 pKa = 4.59TDD594 pKa = 3.27VLTPQDD600 pKa = 3.59TATVPSAPPSSPTPSTNRR618 pKa = 11.84QGEE621 pKa = 4.08AQTQNGEE628 pKa = 4.54DD629 pKa = 4.33SSQDD633 pKa = 2.43WPRR636 pKa = 11.84RR637 pKa = 11.84VKK639 pKa = 9.54TNKK642 pKa = 9.17GRR644 pKa = 11.84EE645 pKa = 4.03FMFPTDD651 pKa = 3.98LLHH654 pKa = 6.23RR655 pKa = 11.84TPPQVLLDD663 pKa = 3.79ALVNEE668 pKa = 4.66YY669 pKa = 10.29EE670 pKa = 4.56SPLSATEE677 pKa = 5.73LSDD680 pKa = 4.09DD681 pKa = 3.6WPEE684 pKa = 3.75MTFEE688 pKa = 3.98EE689 pKa = 4.57RR690 pKa = 11.84KK691 pKa = 10.11NVAFNLL697 pKa = 3.9
Molecular weight: 77.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UVN5|A0A3S8UVN5_9MONO Minor nucleoprotein VP30 OS=Mengla dianlovirus OX=2496529 GN=VP30 PE=1 SV=1
MM1 pKa = 7.78SEE3 pKa = 3.85RR4 pKa = 11.84QSRR7 pKa = 11.84GRR9 pKa = 11.84SRR11 pKa = 11.84TRR13 pKa = 11.84RR14 pKa = 11.84QPDD17 pKa = 3.27EE18 pKa = 4.29VITRR22 pKa = 11.84QEE24 pKa = 3.91VFNPDD29 pKa = 3.17QGPHH33 pKa = 5.53GFHH36 pKa = 6.33RR37 pKa = 11.84RR38 pKa = 11.84SRR40 pKa = 11.84STSNIRR46 pKa = 11.84SNYY49 pKa = 9.0PGGNQYY55 pKa = 10.1TNQIGGGRR63 pKa = 11.84SNQFQPFLRR72 pKa = 11.84PPPPPKK78 pKa = 10.27DD79 pKa = 3.17LCKK82 pKa = 10.33SFKK85 pKa = 10.84VGLPCNDD92 pKa = 3.51KK93 pKa = 11.35GCTKK97 pKa = 10.93DD98 pKa = 4.02HH99 pKa = 7.15DD100 pKa = 4.81LSNLTNRR107 pKa = 11.84EE108 pKa = 4.01LLILIAKK115 pKa = 7.63RR116 pKa = 11.84TFPEE120 pKa = 4.08YY121 pKa = 10.79QKK123 pKa = 10.75IPDD126 pKa = 3.58NTGSKK131 pKa = 10.8GSADD135 pKa = 3.84LSKK138 pKa = 11.13GIPDD142 pKa = 3.69TSNAQNRR149 pKa = 11.84DD150 pKa = 2.88ILTLEE155 pKa = 3.96NLGDD159 pKa = 3.31ILKK162 pKa = 9.95YY163 pKa = 10.84LNSADD168 pKa = 3.94LTTLDD173 pKa = 4.11EE174 pKa = 4.49VSMRR178 pKa = 11.84AALSLTCAGIRR189 pKa = 11.84KK190 pKa = 7.57TSRR193 pKa = 11.84SMINTLTEE201 pKa = 3.78QHH203 pKa = 6.01VSAEE207 pKa = 4.13NLSPDD212 pKa = 2.96QTQIIKK218 pKa = 8.09QTYY221 pKa = 8.21TGIHH225 pKa = 6.66LDD227 pKa = 3.03KK228 pKa = 11.2GGNFEE233 pKa = 4.4AALWKK238 pKa = 10.51NWDD241 pKa = 3.57RR242 pKa = 11.84RR243 pKa = 11.84SISLFLQAAISVLNTTPCEE262 pKa = 4.04SSKK265 pKa = 11.37SVISAYY271 pKa = 10.2NHH273 pKa = 6.44FLQKK277 pKa = 10.58PGDD280 pKa = 3.72IKK282 pKa = 10.7RR283 pKa = 11.84TTLGSINSSVV293 pKa = 2.86
MM1 pKa = 7.78SEE3 pKa = 3.85RR4 pKa = 11.84QSRR7 pKa = 11.84GRR9 pKa = 11.84SRR11 pKa = 11.84TRR13 pKa = 11.84RR14 pKa = 11.84QPDD17 pKa = 3.27EE18 pKa = 4.29VITRR22 pKa = 11.84QEE24 pKa = 3.91VFNPDD29 pKa = 3.17QGPHH33 pKa = 5.53GFHH36 pKa = 6.33RR37 pKa = 11.84RR38 pKa = 11.84SRR40 pKa = 11.84STSNIRR46 pKa = 11.84SNYY49 pKa = 9.0PGGNQYY55 pKa = 10.1TNQIGGGRR63 pKa = 11.84SNQFQPFLRR72 pKa = 11.84PPPPPKK78 pKa = 10.27DD79 pKa = 3.17LCKK82 pKa = 10.33SFKK85 pKa = 10.84VGLPCNDD92 pKa = 3.51KK93 pKa = 11.35GCTKK97 pKa = 10.93DD98 pKa = 4.02HH99 pKa = 7.15DD100 pKa = 4.81LSNLTNRR107 pKa = 11.84EE108 pKa = 4.01LLILIAKK115 pKa = 7.63RR116 pKa = 11.84TFPEE120 pKa = 4.08YY121 pKa = 10.79QKK123 pKa = 10.75IPDD126 pKa = 3.58NTGSKK131 pKa = 10.8GSADD135 pKa = 3.84LSKK138 pKa = 11.13GIPDD142 pKa = 3.69TSNAQNRR149 pKa = 11.84DD150 pKa = 2.88ILTLEE155 pKa = 3.96NLGDD159 pKa = 3.31ILKK162 pKa = 9.95YY163 pKa = 10.84LNSADD168 pKa = 3.94LTTLDD173 pKa = 4.11EE174 pKa = 4.49VSMRR178 pKa = 11.84AALSLTCAGIRR189 pKa = 11.84KK190 pKa = 7.57TSRR193 pKa = 11.84SMINTLTEE201 pKa = 3.78QHH203 pKa = 6.01VSAEE207 pKa = 4.13NLSPDD212 pKa = 2.96QTQIIKK218 pKa = 8.09QTYY221 pKa = 8.21TGIHH225 pKa = 6.66LDD227 pKa = 3.03KK228 pKa = 11.2GGNFEE233 pKa = 4.4AALWKK238 pKa = 10.51NWDD241 pKa = 3.57RR242 pKa = 11.84RR243 pKa = 11.84SISLFLQAAISVLNTTPCEE262 pKa = 4.04SSKK265 pKa = 11.37SVISAYY271 pKa = 10.2NHH273 pKa = 6.44FLQKK277 pKa = 10.58PGDD280 pKa = 3.72IKK282 pKa = 10.7RR283 pKa = 11.84TTLGSINSSVV293 pKa = 2.86
Molecular weight: 32.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4814 |
253 |
2276 |
687.7 |
77.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.629 ± 0.501 | 1.329 ± 0.231 |
4.965 ± 0.504 | 5.526 ± 0.646 |
4.3 ± 0.416 | 5.318 ± 0.473 |
2.327 ± 0.173 | 7.208 ± 0.372 |
6.024 ± 0.351 | 10.656 ± 0.457 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.87 ± 0.213 | 5.526 ± 0.377 |
5.193 ± 0.593 | 5.006 ± 0.479 |
4.882 ± 0.245 | 8.288 ± 0.329 |
6.398 ± 0.479 | 4.965 ± 0.479 |
1.122 ± 0.18 | 3.469 ± 0.672 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
