
Flavobacterium phage FCL-2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Ficleduovirus; Flavobacterium virus FCL2
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
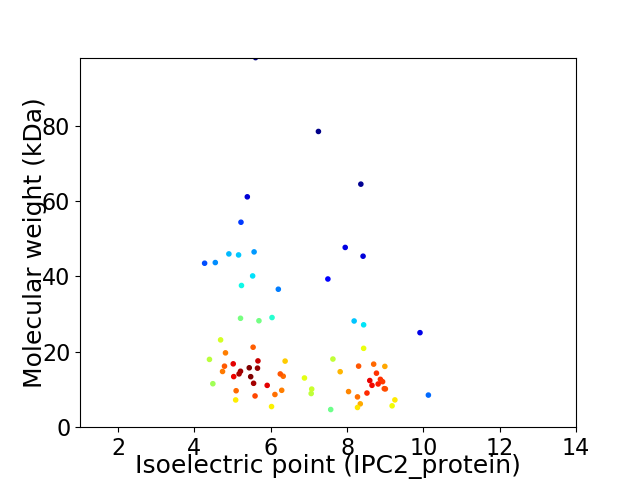
Virtual 2D-PAGE plot for 74 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
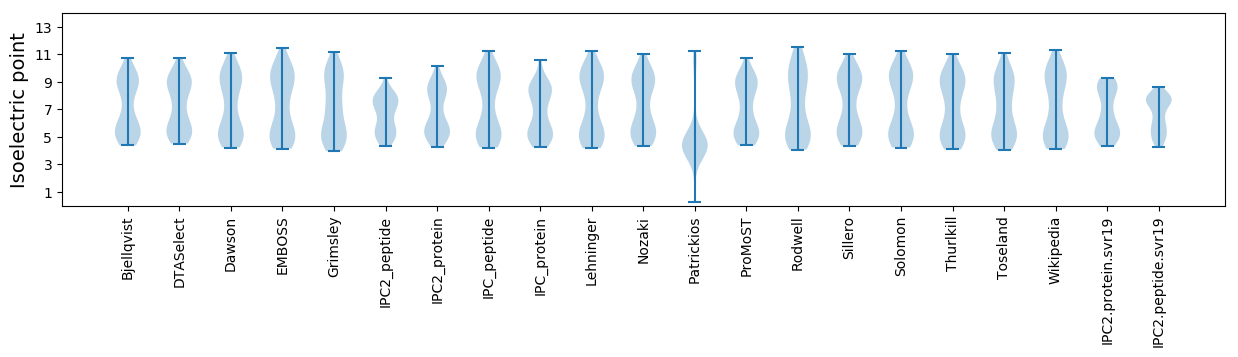
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0YT00|A0A0A0YT00_9CAUD Uncharacterized protein OS=Flavobacterium phage FCL-2 OX=908819 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 10.61VLGNLPIIKK11 pKa = 9.56DD12 pKa = 3.72VNSVYY17 pKa = 10.69PFGAAIKK24 pKa = 9.82NQTEE28 pKa = 4.53TEE30 pKa = 4.02QGTPAIRR37 pKa = 11.84EE38 pKa = 4.08IYY40 pKa = 10.6NDD42 pKa = 3.29VLMNIYY48 pKa = 10.31KK49 pKa = 10.03ILQLTGVNPSNEE61 pKa = 3.68EE62 pKa = 3.85DD63 pKa = 3.45NNYY66 pKa = 10.67NGYY69 pKa = 10.31QIIEE73 pKa = 4.19ALKK76 pKa = 10.49KK77 pKa = 10.72LPNSLNDD84 pKa = 3.17IEE86 pKa = 5.65QILTLDD92 pKa = 3.66GTVWTVPFDD101 pKa = 4.26LQYY104 pKa = 11.47LPNKK108 pKa = 9.86YY109 pKa = 9.25MFVARR114 pKa = 11.84ASEE117 pKa = 4.25NYY119 pKa = 9.99NPAITYY125 pKa = 6.92TFKK128 pKa = 11.25GLGVTEE134 pKa = 5.19LPFNPINGFSATDD147 pKa = 3.52EE148 pKa = 4.28LLVIIDD154 pKa = 3.73TSGVRR159 pKa = 11.84SYY161 pKa = 11.74SLNKK165 pKa = 8.12PTEE168 pKa = 4.14ASSIVYY174 pKa = 8.84TGTGQVVTFNDD185 pKa = 3.92LNTVYY190 pKa = 10.68YY191 pKa = 10.15YY192 pKa = 11.54NNNFSSGWLVADD204 pKa = 4.72TPNSCDD210 pKa = 2.75ILNIVRR216 pKa = 11.84LFASRR221 pKa = 11.84ATLEE225 pKa = 3.88IKK227 pKa = 10.62DD228 pKa = 3.68IFIHH232 pKa = 6.48NNTIVCFCFDD242 pKa = 4.6PSDD245 pKa = 3.53VAYY248 pKa = 10.0FIYY251 pKa = 10.73DD252 pKa = 3.16VDD254 pKa = 4.15LTKK257 pKa = 10.42LTVSFNIEE265 pKa = 3.96LSNDD269 pKa = 2.99ADD271 pKa = 3.91SNYY274 pKa = 10.95DD275 pKa = 3.14PVAYY279 pKa = 9.49MDD281 pKa = 4.07EE282 pKa = 5.11NGDD285 pKa = 2.95LWMTNRR291 pKa = 11.84GNLNGGVNDD300 pKa = 3.46NEE302 pKa = 4.27VYY304 pKa = 10.7KK305 pKa = 10.25FVRR308 pKa = 11.84NGLGIFTFNSTITLGVSFVKK328 pKa = 9.87TRR330 pKa = 11.84NAAIKK335 pKa = 10.33HH336 pKa = 5.27GFLYY340 pKa = 9.7THH342 pKa = 7.19FEE344 pKa = 4.32LYY346 pKa = 10.35LNKK349 pKa = 10.46YY350 pKa = 9.77NLTTGALTSSVYY362 pKa = 10.2FPAIGQLMQLNGEE375 pKa = 4.42VYY377 pKa = 10.63FNSGYY382 pKa = 8.9IAKK385 pKa = 10.25RR386 pKa = 11.84FDD388 pKa = 3.2LL389 pKa = 4.72
MM1 pKa = 7.43KK2 pKa = 10.61VLGNLPIIKK11 pKa = 9.56DD12 pKa = 3.72VNSVYY17 pKa = 10.69PFGAAIKK24 pKa = 9.82NQTEE28 pKa = 4.53TEE30 pKa = 4.02QGTPAIRR37 pKa = 11.84EE38 pKa = 4.08IYY40 pKa = 10.6NDD42 pKa = 3.29VLMNIYY48 pKa = 10.31KK49 pKa = 10.03ILQLTGVNPSNEE61 pKa = 3.68EE62 pKa = 3.85DD63 pKa = 3.45NNYY66 pKa = 10.67NGYY69 pKa = 10.31QIIEE73 pKa = 4.19ALKK76 pKa = 10.49KK77 pKa = 10.72LPNSLNDD84 pKa = 3.17IEE86 pKa = 5.65QILTLDD92 pKa = 3.66GTVWTVPFDD101 pKa = 4.26LQYY104 pKa = 11.47LPNKK108 pKa = 9.86YY109 pKa = 9.25MFVARR114 pKa = 11.84ASEE117 pKa = 4.25NYY119 pKa = 9.99NPAITYY125 pKa = 6.92TFKK128 pKa = 11.25GLGVTEE134 pKa = 5.19LPFNPINGFSATDD147 pKa = 3.52EE148 pKa = 4.28LLVIIDD154 pKa = 3.73TSGVRR159 pKa = 11.84SYY161 pKa = 11.74SLNKK165 pKa = 8.12PTEE168 pKa = 4.14ASSIVYY174 pKa = 8.84TGTGQVVTFNDD185 pKa = 3.92LNTVYY190 pKa = 10.68YY191 pKa = 10.15YY192 pKa = 11.54NNNFSSGWLVADD204 pKa = 4.72TPNSCDD210 pKa = 2.75ILNIVRR216 pKa = 11.84LFASRR221 pKa = 11.84ATLEE225 pKa = 3.88IKK227 pKa = 10.62DD228 pKa = 3.68IFIHH232 pKa = 6.48NNTIVCFCFDD242 pKa = 4.6PSDD245 pKa = 3.53VAYY248 pKa = 10.0FIYY251 pKa = 10.73DD252 pKa = 3.16VDD254 pKa = 4.15LTKK257 pKa = 10.42LTVSFNIEE265 pKa = 3.96LSNDD269 pKa = 2.99ADD271 pKa = 3.91SNYY274 pKa = 10.95DD275 pKa = 3.14PVAYY279 pKa = 9.49MDD281 pKa = 4.07EE282 pKa = 5.11NGDD285 pKa = 2.95LWMTNRR291 pKa = 11.84GNLNGGVNDD300 pKa = 3.46NEE302 pKa = 4.27VYY304 pKa = 10.7KK305 pKa = 10.25FVRR308 pKa = 11.84NGLGIFTFNSTITLGVSFVKK328 pKa = 9.87TRR330 pKa = 11.84NAAIKK335 pKa = 10.33HH336 pKa = 5.27GFLYY340 pKa = 9.7THH342 pKa = 7.19FEE344 pKa = 4.32LYY346 pKa = 10.35LNKK349 pKa = 10.46YY350 pKa = 9.77NLTTGALTSSVYY362 pKa = 10.2FPAIGQLMQLNGEE375 pKa = 4.42VYY377 pKa = 10.63FNSGYY382 pKa = 8.9IAKK385 pKa = 10.25RR386 pKa = 11.84FDD388 pKa = 3.2LL389 pKa = 4.72
Molecular weight: 43.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0YQ72|A0A0A0YQ72_9CAUD Uncharacterized protein OS=Flavobacterium phage FCL-2 OX=908819 PE=4 SV=1
MM1 pKa = 7.56RR2 pKa = 11.84AWNANLRR9 pKa = 11.84LKK11 pKa = 10.72ANRR14 pKa = 11.84VVYY17 pKa = 10.17RR18 pKa = 11.84AYY20 pKa = 8.46THH22 pKa = 6.43NMVKK26 pKa = 8.85PTGTTDD32 pKa = 3.4YY33 pKa = 10.89VSVSFVLPVKK43 pKa = 10.66SDD45 pKa = 3.41FLANGGLLYY54 pKa = 10.87VNPNTGITITNRR66 pKa = 11.84YY67 pKa = 7.56IAAGLLHH74 pKa = 6.79KK75 pKa = 10.68GVV77 pKa = 3.71
MM1 pKa = 7.56RR2 pKa = 11.84AWNANLRR9 pKa = 11.84LKK11 pKa = 10.72ANRR14 pKa = 11.84VVYY17 pKa = 10.17RR18 pKa = 11.84AYY20 pKa = 8.46THH22 pKa = 6.43NMVKK26 pKa = 8.85PTGTTDD32 pKa = 3.4YY33 pKa = 10.89VSVSFVLPVKK43 pKa = 10.66SDD45 pKa = 3.41FLANGGLLYY54 pKa = 10.87VNPNTGITITNRR66 pKa = 11.84YY67 pKa = 7.56IAAGLLHH74 pKa = 6.79KK75 pKa = 10.68GVV77 pKa = 3.71
Molecular weight: 8.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14454 |
41 |
841 |
195.3 |
22.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.237 ± 0.459 | 0.941 ± 0.116 |
5.825 ± 0.247 | 7.23 ± 0.452 |
4.725 ± 0.175 | 5.521 ± 0.293 |
1.335 ± 0.128 | 7.991 ± 0.256 |
9.202 ± 0.406 | 8.655 ± 0.22 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.235 ± 0.159 | 7.292 ± 0.298 |
2.927 ± 0.161 | 3.667 ± 0.192 |
3.722 ± 0.206 | 6.337 ± 0.281 |
5.978 ± 0.248 | 5.825 ± 0.231 |
1.072 ± 0.127 | 4.283 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
