
Porcine feces-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus anima1
Average proteome isoelectric point is 8.75
Get precalculated fractions of proteins
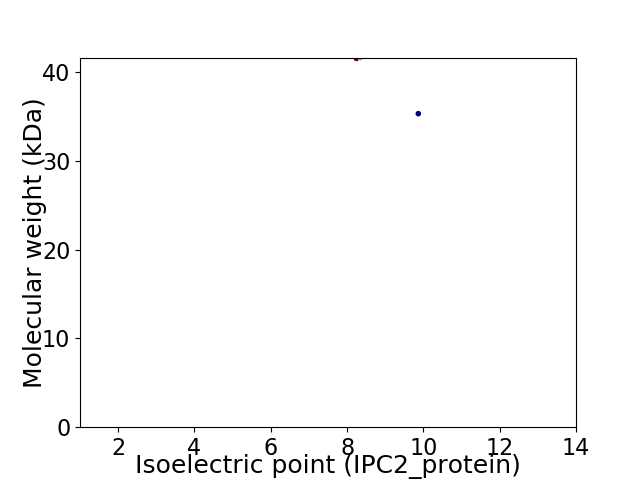
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
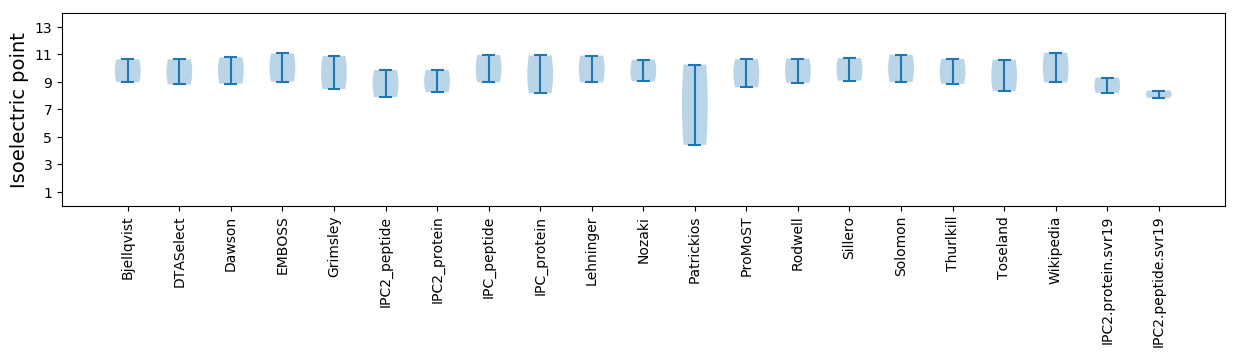
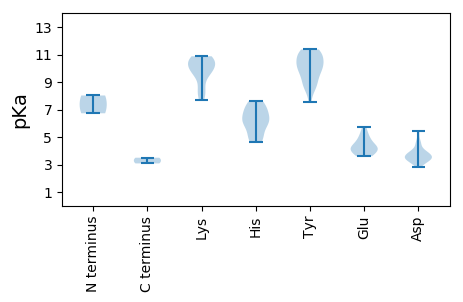
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A221LEA2|A0A221LEA2_9VIRU Capsid OS=Porcine feces-associated gemycircularvirus OX=2017712 PE=4 SV=1
MM1 pKa = 6.73VQYY4 pKa = 10.71NVNSWTIRR12 pKa = 11.84PFRR15 pKa = 11.84KK16 pKa = 9.8LMSSFRR22 pKa = 11.84FQARR26 pKa = 11.84YY27 pKa = 10.11GLFTYY32 pKa = 7.58SQCGRR37 pKa = 11.84LRR39 pKa = 11.84PQQVSEE45 pKa = 4.48HH46 pKa = 5.12FTSLGADD53 pKa = 3.66HH54 pKa = 7.29IIGRR58 pKa = 11.84EE59 pKa = 3.79THH61 pKa = 5.99QDD63 pKa = 3.12GGTHH67 pKa = 5.9LHH69 pKa = 5.88VFADD73 pKa = 4.56FRR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.94YY78 pKa = 9.02RR79 pKa = 11.84TRR81 pKa = 11.84DD82 pKa = 3.02NRR84 pKa = 11.84KK85 pKa = 8.04FDD87 pKa = 3.6VEE89 pKa = 4.33GFHH92 pKa = 6.93PNIVPSLRR100 pKa = 11.84NPAGGWDD107 pKa = 3.84YY108 pKa = 10.92ATKK111 pKa = 10.83DD112 pKa = 3.2GDD114 pKa = 3.43ICAGEE119 pKa = 4.27LSRR122 pKa = 11.84PDD124 pKa = 3.67DD125 pKa = 3.77EE126 pKa = 5.41RR127 pKa = 11.84GSRR130 pKa = 11.84SRR132 pKa = 11.84AADD135 pKa = 3.74AGWDD139 pKa = 3.53ALRR142 pKa = 11.84AAEE145 pKa = 4.19TRR147 pKa = 11.84DD148 pKa = 3.33DD149 pKa = 3.96FFRR152 pKa = 11.84IASEE156 pKa = 4.06HH157 pKa = 6.19LFSHH161 pKa = 6.53LVKK164 pKa = 10.71SFNSFSAYY172 pKa = 10.37ADD174 pKa = 2.82WKK176 pKa = 10.51FRR178 pKa = 11.84VDD180 pKa = 3.55RR181 pKa = 11.84TPYY184 pKa = 9.52IHH186 pKa = 7.59DD187 pKa = 4.32PNVTFDD193 pKa = 4.76LGQLHH198 pKa = 6.29QLRR201 pKa = 11.84EE202 pKa = 4.47WIRR205 pKa = 11.84GNLPPGNSKK214 pKa = 10.32YY215 pKa = 9.04RR216 pKa = 11.84TLHH219 pKa = 6.14LRR221 pKa = 11.84VQSIEE226 pKa = 3.65LRR228 pKa = 11.84EE229 pKa = 3.99RR230 pKa = 11.84GISLLLWGRR239 pKa = 11.84SRR241 pKa = 11.84TGKK244 pKa = 7.69TWLARR249 pKa = 11.84SFGSHH254 pKa = 6.83CYY256 pKa = 10.22FGGLFALDD264 pKa = 3.44EE265 pKa = 4.33YY266 pKa = 11.04EE267 pKa = 4.67YY268 pKa = 11.33NDD270 pKa = 3.63KK271 pKa = 10.87VDD273 pKa = 3.48YY274 pKa = 11.18AVFDD278 pKa = 5.21DD279 pKa = 3.75IQGGFKK285 pKa = 10.57YY286 pKa = 10.42FPAYY290 pKa = 9.71KK291 pKa = 9.97SWLGQQAEE299 pKa = 5.02FYY301 pKa = 9.88CTDD304 pKa = 3.05KK305 pKa = 10.79FRR307 pKa = 11.84KK308 pKa = 9.1KK309 pKa = 10.52KK310 pKa = 8.41YY311 pKa = 9.24IKK313 pKa = 9.54WGRR316 pKa = 11.84PCIWIMNEE324 pKa = 3.73DD325 pKa = 4.63PYY327 pKa = 10.22TQEE330 pKa = 5.17VDD332 pKa = 3.45IDD334 pKa = 3.78WLEE337 pKa = 4.06KK338 pKa = 10.45NCLIVHH344 pKa = 7.01IDD346 pKa = 3.45RR347 pKa = 11.84PLYY350 pKa = 8.64QTAA353 pKa = 3.5
MM1 pKa = 6.73VQYY4 pKa = 10.71NVNSWTIRR12 pKa = 11.84PFRR15 pKa = 11.84KK16 pKa = 9.8LMSSFRR22 pKa = 11.84FQARR26 pKa = 11.84YY27 pKa = 10.11GLFTYY32 pKa = 7.58SQCGRR37 pKa = 11.84LRR39 pKa = 11.84PQQVSEE45 pKa = 4.48HH46 pKa = 5.12FTSLGADD53 pKa = 3.66HH54 pKa = 7.29IIGRR58 pKa = 11.84EE59 pKa = 3.79THH61 pKa = 5.99QDD63 pKa = 3.12GGTHH67 pKa = 5.9LHH69 pKa = 5.88VFADD73 pKa = 4.56FRR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.94YY78 pKa = 9.02RR79 pKa = 11.84TRR81 pKa = 11.84DD82 pKa = 3.02NRR84 pKa = 11.84KK85 pKa = 8.04FDD87 pKa = 3.6VEE89 pKa = 4.33GFHH92 pKa = 6.93PNIVPSLRR100 pKa = 11.84NPAGGWDD107 pKa = 3.84YY108 pKa = 10.92ATKK111 pKa = 10.83DD112 pKa = 3.2GDD114 pKa = 3.43ICAGEE119 pKa = 4.27LSRR122 pKa = 11.84PDD124 pKa = 3.67DD125 pKa = 3.77EE126 pKa = 5.41RR127 pKa = 11.84GSRR130 pKa = 11.84SRR132 pKa = 11.84AADD135 pKa = 3.74AGWDD139 pKa = 3.53ALRR142 pKa = 11.84AAEE145 pKa = 4.19TRR147 pKa = 11.84DD148 pKa = 3.33DD149 pKa = 3.96FFRR152 pKa = 11.84IASEE156 pKa = 4.06HH157 pKa = 6.19LFSHH161 pKa = 6.53LVKK164 pKa = 10.71SFNSFSAYY172 pKa = 10.37ADD174 pKa = 2.82WKK176 pKa = 10.51FRR178 pKa = 11.84VDD180 pKa = 3.55RR181 pKa = 11.84TPYY184 pKa = 9.52IHH186 pKa = 7.59DD187 pKa = 4.32PNVTFDD193 pKa = 4.76LGQLHH198 pKa = 6.29QLRR201 pKa = 11.84EE202 pKa = 4.47WIRR205 pKa = 11.84GNLPPGNSKK214 pKa = 10.32YY215 pKa = 9.04RR216 pKa = 11.84TLHH219 pKa = 6.14LRR221 pKa = 11.84VQSIEE226 pKa = 3.65LRR228 pKa = 11.84EE229 pKa = 3.99RR230 pKa = 11.84GISLLLWGRR239 pKa = 11.84SRR241 pKa = 11.84TGKK244 pKa = 7.69TWLARR249 pKa = 11.84SFGSHH254 pKa = 6.83CYY256 pKa = 10.22FGGLFALDD264 pKa = 3.44EE265 pKa = 4.33YY266 pKa = 11.04EE267 pKa = 4.67YY268 pKa = 11.33NDD270 pKa = 3.63KK271 pKa = 10.87VDD273 pKa = 3.48YY274 pKa = 11.18AVFDD278 pKa = 5.21DD279 pKa = 3.75IQGGFKK285 pKa = 10.57YY286 pKa = 10.42FPAYY290 pKa = 9.71KK291 pKa = 9.97SWLGQQAEE299 pKa = 5.02FYY301 pKa = 9.88CTDD304 pKa = 3.05KK305 pKa = 10.79FRR307 pKa = 11.84KK308 pKa = 9.1KK309 pKa = 10.52KK310 pKa = 8.41YY311 pKa = 9.24IKK313 pKa = 9.54WGRR316 pKa = 11.84PCIWIMNEE324 pKa = 3.73DD325 pKa = 4.63PYY327 pKa = 10.22TQEE330 pKa = 5.17VDD332 pKa = 3.45IDD334 pKa = 3.78WLEE337 pKa = 4.06KK338 pKa = 10.45NCLIVHH344 pKa = 7.01IDD346 pKa = 3.45RR347 pKa = 11.84PLYY350 pKa = 8.64QTAA353 pKa = 3.5
Molecular weight: 41.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A221LEA2|A0A221LEA2_9VIRU Capsid OS=Porcine feces-associated gemycircularvirus OX=2017712 PE=4 SV=1
MM1 pKa = 8.02AYY3 pKa = 10.14RR4 pKa = 11.84FRR6 pKa = 11.84RR7 pKa = 11.84FSTRR11 pKa = 11.84SRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FTPRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84SSFRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84NYY29 pKa = 10.62RR30 pKa = 11.84MMRR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84RR37 pKa = 11.84FSRR40 pKa = 11.84PRR42 pKa = 11.84TNVRR46 pKa = 11.84RR47 pKa = 11.84VRR49 pKa = 11.84NLASRR54 pKa = 11.84KK55 pKa = 9.61CRR57 pKa = 11.84DD58 pKa = 3.33NMLSNPIDD66 pKa = 3.78DD67 pKa = 4.48TGSPTTPGPVTMTGGNTYY85 pKa = 11.14AFIFSPSARR94 pKa = 11.84VAGNEE99 pKa = 3.61ISSGDD104 pKa = 3.56RR105 pKa = 11.84VNQPSSSQRR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.89SKK118 pKa = 10.77CFVSGYY124 pKa = 10.48GEE126 pKa = 4.14TVEE129 pKa = 4.76IQTDD133 pKa = 3.94DD134 pKa = 3.29ASPWVWRR141 pKa = 11.84RR142 pKa = 11.84IVFSTIGLAVQFQPGLLYY160 pKa = 11.26NNDD163 pKa = 3.23DD164 pKa = 3.33TRR166 pKa = 11.84GYY168 pKa = 11.15EE169 pKa = 3.61RR170 pKa = 11.84TTFNLASGTAQADD183 pKa = 3.51QLQGIVYY190 pKa = 9.62RR191 pKa = 11.84YY192 pKa = 9.63LFQGSQDD199 pKa = 3.39IDD201 pKa = 3.3WANVMTANTAPRR213 pKa = 11.84HH214 pKa = 5.01VKK216 pKa = 9.97VYY218 pKa = 8.6PDD220 pKa = 3.47RR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84VIQSPNSEE231 pKa = 3.95GGTIRR236 pKa = 11.84ISKK239 pKa = 10.06HH240 pKa = 4.66YY241 pKa = 10.81DD242 pKa = 2.98NIRR245 pKa = 11.84RR246 pKa = 11.84SIIYY250 pKa = 10.02DD251 pKa = 4.07DD252 pKa = 4.12IEE254 pKa = 5.7AGSGAKK260 pKa = 8.39ATNAIASGTPYY271 pKa = 11.37GNMGDD276 pKa = 3.97LFVVDD281 pKa = 5.42FFSSASDD288 pKa = 3.96DD289 pKa = 3.79EE290 pKa = 4.86TTGATFQPHH299 pKa = 4.82GRR301 pKa = 11.84YY302 pKa = 8.68YY303 pKa = 8.44WHH305 pKa = 7.46EE306 pKa = 4.17GQAVV310 pKa = 3.08
MM1 pKa = 8.02AYY3 pKa = 10.14RR4 pKa = 11.84FRR6 pKa = 11.84RR7 pKa = 11.84FSTRR11 pKa = 11.84SRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FTPRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84SSFRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84NYY29 pKa = 10.62RR30 pKa = 11.84MMRR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84RR37 pKa = 11.84FSRR40 pKa = 11.84PRR42 pKa = 11.84TNVRR46 pKa = 11.84RR47 pKa = 11.84VRR49 pKa = 11.84NLASRR54 pKa = 11.84KK55 pKa = 9.61CRR57 pKa = 11.84DD58 pKa = 3.33NMLSNPIDD66 pKa = 3.78DD67 pKa = 4.48TGSPTTPGPVTMTGGNTYY85 pKa = 11.14AFIFSPSARR94 pKa = 11.84VAGNEE99 pKa = 3.61ISSGDD104 pKa = 3.56RR105 pKa = 11.84VNQPSSSQRR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.89SKK118 pKa = 10.77CFVSGYY124 pKa = 10.48GEE126 pKa = 4.14TVEE129 pKa = 4.76IQTDD133 pKa = 3.94DD134 pKa = 3.29ASPWVWRR141 pKa = 11.84RR142 pKa = 11.84IVFSTIGLAVQFQPGLLYY160 pKa = 11.26NNDD163 pKa = 3.23DD164 pKa = 3.33TRR166 pKa = 11.84GYY168 pKa = 11.15EE169 pKa = 3.61RR170 pKa = 11.84TTFNLASGTAQADD183 pKa = 3.51QLQGIVYY190 pKa = 9.62RR191 pKa = 11.84YY192 pKa = 9.63LFQGSQDD199 pKa = 3.39IDD201 pKa = 3.3WANVMTANTAPRR213 pKa = 11.84HH214 pKa = 5.01VKK216 pKa = 9.97VYY218 pKa = 8.6PDD220 pKa = 3.47RR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84VIQSPNSEE231 pKa = 3.95GGTIRR236 pKa = 11.84ISKK239 pKa = 10.06HH240 pKa = 4.66YY241 pKa = 10.81DD242 pKa = 2.98NIRR245 pKa = 11.84RR246 pKa = 11.84SIIYY250 pKa = 10.02DD251 pKa = 4.07DD252 pKa = 4.12IEE254 pKa = 5.7AGSGAKK260 pKa = 8.39ATNAIASGTPYY271 pKa = 11.37GNMGDD276 pKa = 3.97LFVVDD281 pKa = 5.42FFSSASDD288 pKa = 3.96DD289 pKa = 3.79EE290 pKa = 4.86TTGATFQPHH299 pKa = 4.82GRR301 pKa = 11.84YY302 pKa = 8.68YY303 pKa = 8.44WHH305 pKa = 7.46EE306 pKa = 4.17GQAVV310 pKa = 3.08
Molecular weight: 35.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
663 |
310 |
353 |
331.5 |
38.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.335 ± 0.286 | 1.207 ± 0.366 |
7.089 ± 0.625 | 3.62 ± 0.677 |
6.033 ± 0.568 | 7.541 ± 0.131 |
2.564 ± 0.83 | 4.977 ± 0.12 |
3.469 ± 0.999 | 5.43 ± 1.646 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.508 ± 0.488 | 4.374 ± 0.723 |
4.525 ± 0.415 | 4.072 ± 0.079 |
11.463 ± 1.358 | 7.843 ± 1.195 |
6.033 ± 1.113 | 4.827 ± 0.638 |
2.262 ± 0.633 | 4.827 ± 0.202 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
