
ANME-2 cluster archaeon
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; unclassified Methanosarcinales; ANME-2 cluster; unclassified ANME-2 cluster
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
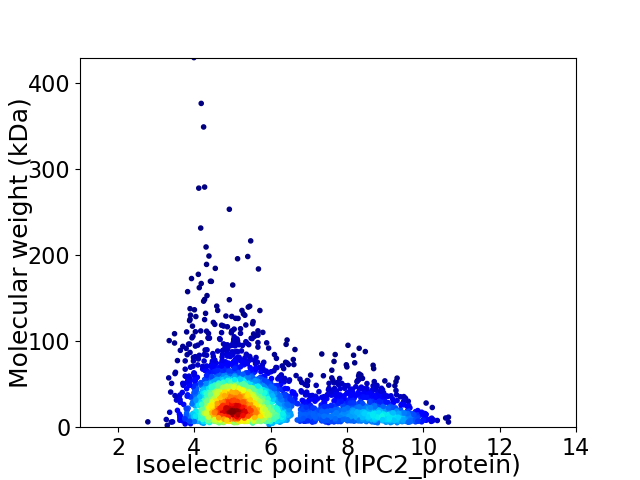
Virtual 2D-PAGE plot for 3377 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V3JK96|A0A2V3JK96_9EURY FmdE domain-containing protein OS=ANME-2 cluster archaeon OX=2056317 GN=C4B59_07205 PE=4 SV=1
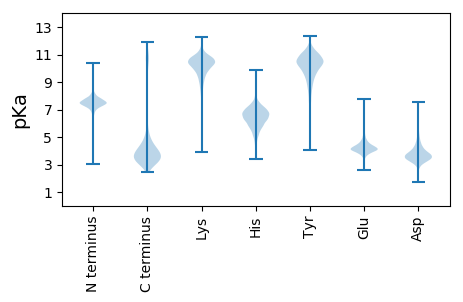
MM1 pKa = 7.22TRR3 pKa = 11.84LPLVAAAVVILAFAGVGAASMPPVPYY29 pKa = 10.42FSQCDD34 pKa = 3.8PRR36 pKa = 11.84WGSDD40 pKa = 2.9KK41 pKa = 11.28LGGDD45 pKa = 3.44GQTIYY50 pKa = 11.09SQGCTLTSAAMVMAYY65 pKa = 10.3YY66 pKa = 10.62GVDD69 pKa = 3.48TDD71 pKa = 4.42PKK73 pKa = 10.19RR74 pKa = 11.84LNDD77 pKa = 4.35AIGRR81 pKa = 11.84AGYY84 pKa = 10.02DD85 pKa = 3.23EE86 pKa = 5.4NYY88 pKa = 8.33WIHH91 pKa = 6.42WSAVSDD97 pKa = 3.67ACHH100 pKa = 7.33DD101 pKa = 3.54EE102 pKa = 4.38TNQIEE107 pKa = 4.45YY108 pKa = 10.91SPGTVKK114 pKa = 10.68PFDD117 pKa = 3.61TTVLNTHH124 pKa = 7.2LDD126 pKa = 3.35AGHH129 pKa = 6.42PVIVNVGGHH138 pKa = 5.18FVVVTGRR145 pKa = 11.84SDD147 pKa = 2.94GTYY150 pKa = 10.24YY151 pKa = 10.36INDD154 pKa = 5.12PISSAKK160 pKa = 8.97STLDD164 pKa = 3.21AYY166 pKa = 9.58PNRR169 pKa = 11.84VGMHH173 pKa = 6.74IYY175 pKa = 10.32SGNPPDD181 pKa = 3.54INKK184 pKa = 9.58PLVGDD189 pKa = 3.64WNGNGTDD196 pKa = 3.15TTGIYY201 pKa = 10.54NYY203 pKa = 7.5TTANFSLDD211 pKa = 3.22SGLNISFSISGDD223 pKa = 3.27IPITGDD229 pKa = 2.97WNGNGYY235 pKa = 8.17DD236 pKa = 3.69TIGVFRR242 pKa = 11.84PSTAQFFLDD251 pKa = 3.74YY252 pKa = 11.22DD253 pKa = 3.85NDD255 pKa = 4.18GVSDD259 pKa = 3.6MNATFGVIGDD269 pKa = 3.92IPVVGDD275 pKa = 3.27WDD277 pKa = 3.98GDD279 pKa = 3.67GGDD282 pKa = 3.74NIGVFRR288 pKa = 11.84QNHH291 pKa = 5.82SDD293 pKa = 3.16SGLTMFFLDD302 pKa = 4.74LDD304 pKa = 4.07NSGGSADD311 pKa = 3.45QSVAFGEE318 pKa = 4.37PDD320 pKa = 3.95DD321 pKa = 5.42LPVIGDD327 pKa = 3.49WDD329 pKa = 3.94GDD331 pKa = 3.63GDD333 pKa = 5.03DD334 pKa = 5.29NIGLYY339 pKa = 10.36RR340 pKa = 11.84PGSTTGVFYY349 pKa = 11.25LDD351 pKa = 3.69IDD353 pKa = 4.13NDD355 pKa = 3.63GGAADD360 pKa = 4.22IVTPEE365 pKa = 4.1YY366 pKa = 11.15GDD368 pKa = 4.67LGDD371 pKa = 5.1IPIAGDD377 pKa = 3.29WDD379 pKa = 3.9GDD381 pKa = 3.71SDD383 pKa = 5.38DD384 pKa = 5.04NIGVYY389 pKa = 10.07RR390 pKa = 11.84PGTGEE395 pKa = 3.92FFLNASMPSVPKK407 pKa = 10.06TIYY410 pKa = 10.76VDD412 pKa = 5.16DD413 pKa = 5.42DD414 pKa = 4.56FSDD417 pKa = 4.47DD418 pKa = 3.9PSEE421 pKa = 4.72HH422 pKa = 6.54KK423 pKa = 10.02WDD425 pKa = 4.46TIQEE429 pKa = 4.41GVDD432 pKa = 3.51DD433 pKa = 4.58ATDD436 pKa = 3.69GDD438 pKa = 4.52TVIVYY443 pKa = 10.28AGEE446 pKa = 4.04YY447 pKa = 10.07NEE449 pKa = 4.37TVGVGVEE456 pKa = 4.02NSITLWGAGVDD467 pKa = 3.94VVTVRR472 pKa = 11.84SKK474 pKa = 9.8CTWGRR479 pKa = 11.84VFDD482 pKa = 3.79VTAGYY487 pKa = 7.6TRR489 pKa = 11.84HH490 pKa = 4.94RR491 pKa = 11.84HH492 pKa = 4.83KK493 pKa = 11.12LRR495 pKa = 11.84FFYY498 pKa = 10.63RR499 pKa = 3.45
MM1 pKa = 7.22TRR3 pKa = 11.84LPLVAAAVVILAFAGVGAASMPPVPYY29 pKa = 10.42FSQCDD34 pKa = 3.8PRR36 pKa = 11.84WGSDD40 pKa = 2.9KK41 pKa = 11.28LGGDD45 pKa = 3.44GQTIYY50 pKa = 11.09SQGCTLTSAAMVMAYY65 pKa = 10.3YY66 pKa = 10.62GVDD69 pKa = 3.48TDD71 pKa = 4.42PKK73 pKa = 10.19RR74 pKa = 11.84LNDD77 pKa = 4.35AIGRR81 pKa = 11.84AGYY84 pKa = 10.02DD85 pKa = 3.23EE86 pKa = 5.4NYY88 pKa = 8.33WIHH91 pKa = 6.42WSAVSDD97 pKa = 3.67ACHH100 pKa = 7.33DD101 pKa = 3.54EE102 pKa = 4.38TNQIEE107 pKa = 4.45YY108 pKa = 10.91SPGTVKK114 pKa = 10.68PFDD117 pKa = 3.61TTVLNTHH124 pKa = 7.2LDD126 pKa = 3.35AGHH129 pKa = 6.42PVIVNVGGHH138 pKa = 5.18FVVVTGRR145 pKa = 11.84SDD147 pKa = 2.94GTYY150 pKa = 10.24YY151 pKa = 10.36INDD154 pKa = 5.12PISSAKK160 pKa = 8.97STLDD164 pKa = 3.21AYY166 pKa = 9.58PNRR169 pKa = 11.84VGMHH173 pKa = 6.74IYY175 pKa = 10.32SGNPPDD181 pKa = 3.54INKK184 pKa = 9.58PLVGDD189 pKa = 3.64WNGNGTDD196 pKa = 3.15TTGIYY201 pKa = 10.54NYY203 pKa = 7.5TTANFSLDD211 pKa = 3.22SGLNISFSISGDD223 pKa = 3.27IPITGDD229 pKa = 2.97WNGNGYY235 pKa = 8.17DD236 pKa = 3.69TIGVFRR242 pKa = 11.84PSTAQFFLDD251 pKa = 3.74YY252 pKa = 11.22DD253 pKa = 3.85NDD255 pKa = 4.18GVSDD259 pKa = 3.6MNATFGVIGDD269 pKa = 3.92IPVVGDD275 pKa = 3.27WDD277 pKa = 3.98GDD279 pKa = 3.67GGDD282 pKa = 3.74NIGVFRR288 pKa = 11.84QNHH291 pKa = 5.82SDD293 pKa = 3.16SGLTMFFLDD302 pKa = 4.74LDD304 pKa = 4.07NSGGSADD311 pKa = 3.45QSVAFGEE318 pKa = 4.37PDD320 pKa = 3.95DD321 pKa = 5.42LPVIGDD327 pKa = 3.49WDD329 pKa = 3.94GDD331 pKa = 3.63GDD333 pKa = 5.03DD334 pKa = 5.29NIGLYY339 pKa = 10.36RR340 pKa = 11.84PGSTTGVFYY349 pKa = 11.25LDD351 pKa = 3.69IDD353 pKa = 4.13NDD355 pKa = 3.63GGAADD360 pKa = 4.22IVTPEE365 pKa = 4.1YY366 pKa = 11.15GDD368 pKa = 4.67LGDD371 pKa = 5.1IPIAGDD377 pKa = 3.29WDD379 pKa = 3.9GDD381 pKa = 3.71SDD383 pKa = 5.38DD384 pKa = 5.04NIGVYY389 pKa = 10.07RR390 pKa = 11.84PGTGEE395 pKa = 3.92FFLNASMPSVPKK407 pKa = 10.06TIYY410 pKa = 10.76VDD412 pKa = 5.16DD413 pKa = 5.42DD414 pKa = 4.56FSDD417 pKa = 4.47DD418 pKa = 3.9PSEE421 pKa = 4.72HH422 pKa = 6.54KK423 pKa = 10.02WDD425 pKa = 4.46TIQEE429 pKa = 4.41GVDD432 pKa = 3.51DD433 pKa = 4.58ATDD436 pKa = 3.69GDD438 pKa = 4.52TVIVYY443 pKa = 10.28AGEE446 pKa = 4.04YY447 pKa = 10.07NEE449 pKa = 4.37TVGVGVEE456 pKa = 4.02NSITLWGAGVDD467 pKa = 3.94VVTVRR472 pKa = 11.84SKK474 pKa = 9.8CTWGRR479 pKa = 11.84VFDD482 pKa = 3.79VTAGYY487 pKa = 7.6TRR489 pKa = 11.84HH490 pKa = 4.94RR491 pKa = 11.84HH492 pKa = 4.83KK493 pKa = 11.12LRR495 pKa = 11.84FFYY498 pKa = 10.63RR499 pKa = 3.45
Molecular weight: 53.63 kDa
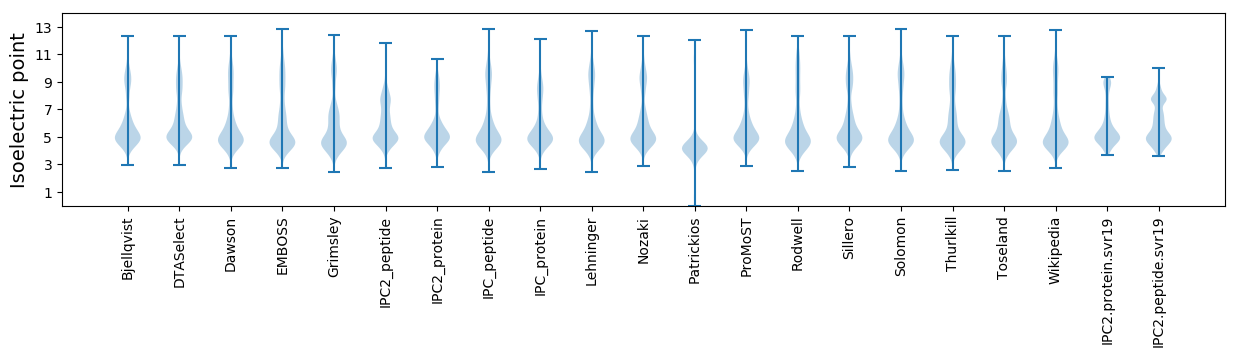
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V3JI79|A0A2V3JI79_9EURY Uncharacterized protein OS=ANME-2 cluster archaeon OX=2056317 GN=C4B59_11875 PE=4 SV=1
MM1 pKa = 7.21GQIEE5 pKa = 4.73VQNLPQPEE13 pKa = 4.09MVLTYY18 pKa = 10.35RR19 pKa = 11.84AVFHH23 pKa = 6.63CMHH26 pKa = 7.03GGKK29 pKa = 9.5SVNVNSSNTPLKK41 pKa = 11.14GEE43 pKa = 3.87ILTNIRR49 pKa = 11.84PKK51 pKa = 11.11LNWTDD56 pKa = 3.5INWVKK61 pKa = 10.76VEE63 pKa = 3.95EE64 pKa = 4.27HH65 pKa = 6.61ANRR68 pKa = 11.84LQIRR72 pKa = 11.84ITMAVKK78 pKa = 9.83TEE80 pKa = 4.01INSCPGTEE88 pKa = 3.51RR89 pKa = 11.84VTRR92 pKa = 11.84CLTAKK97 pKa = 9.04QRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84QRR103 pKa = 11.84YY104 pKa = 7.65EE105 pKa = 3.71GKK107 pKa = 10.46LSCTVLRR114 pKa = 11.84RR115 pKa = 11.84GEE117 pKa = 4.01AGNRR121 pKa = 11.84LVLSRR126 pKa = 11.84RR127 pKa = 11.84LKK129 pKa = 10.02MLL131 pKa = 3.86
MM1 pKa = 7.21GQIEE5 pKa = 4.73VQNLPQPEE13 pKa = 4.09MVLTYY18 pKa = 10.35RR19 pKa = 11.84AVFHH23 pKa = 6.63CMHH26 pKa = 7.03GGKK29 pKa = 9.5SVNVNSSNTPLKK41 pKa = 11.14GEE43 pKa = 3.87ILTNIRR49 pKa = 11.84PKK51 pKa = 11.11LNWTDD56 pKa = 3.5INWVKK61 pKa = 10.76VEE63 pKa = 3.95EE64 pKa = 4.27HH65 pKa = 6.61ANRR68 pKa = 11.84LQIRR72 pKa = 11.84ITMAVKK78 pKa = 9.83TEE80 pKa = 4.01INSCPGTEE88 pKa = 3.51RR89 pKa = 11.84VTRR92 pKa = 11.84CLTAKK97 pKa = 9.04QRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84QRR103 pKa = 11.84YY104 pKa = 7.65EE105 pKa = 3.71GKK107 pKa = 10.46LSCTVLRR114 pKa = 11.84RR115 pKa = 11.84GEE117 pKa = 4.01AGNRR121 pKa = 11.84LVLSRR126 pKa = 11.84RR127 pKa = 11.84LKK129 pKa = 10.02MLL131 pKa = 3.86
Molecular weight: 15.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
955486 |
22 |
3892 |
282.9 |
31.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.584 ± 0.058 | 1.423 ± 0.023 |
6.705 ± 0.045 | 6.985 ± 0.043 |
3.672 ± 0.033 | 7.716 ± 0.047 |
2.06 ± 0.021 | 7.707 ± 0.045 |
5.069 ± 0.046 | 8.621 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.025 | 3.988 ± 0.066 |
4.044 ± 0.028 | 2.342 ± 0.023 |
5.331 ± 0.045 | 6.232 ± 0.036 |
5.875 ± 0.047 | 7.384 ± 0.041 |
1.144 ± 0.02 | 3.485 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
