
Mumps virus (strain Miyahara vaccine) (MuV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Orthorubulavirus; Mumps orthorubulavirus
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
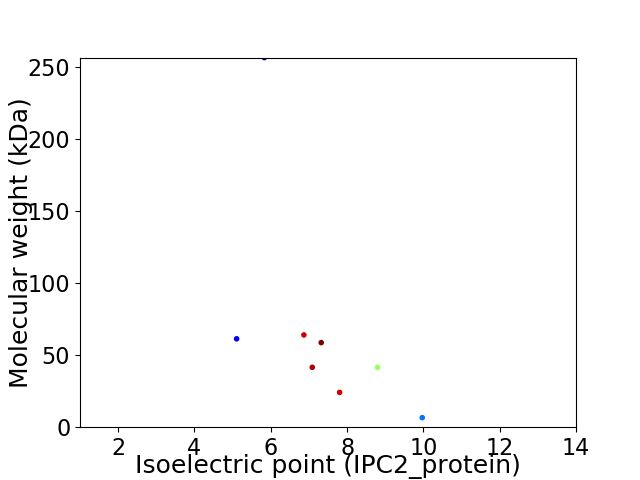
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|P22112|SH_MUMPM Small hydrophobic protein OS=Mumps virus (strain Miyahara vaccine) OX=11171 GN=SH PE=1 SV=1
MM1 pKa = 7.49SSVLKK6 pKa = 10.65AFEE9 pKa = 4.31RR10 pKa = 11.84FTIEE14 pKa = 5.13QEE16 pKa = 3.95LQDD19 pKa = 3.9RR20 pKa = 11.84GEE22 pKa = 4.45EE23 pKa = 4.26GSIPPEE29 pKa = 4.03TLKK32 pKa = 11.08SAVKK36 pKa = 10.23VFVINTPNPTTRR48 pKa = 11.84YY49 pKa = 10.18HH50 pKa = 6.21MLNFCLRR57 pKa = 11.84IICSQNARR65 pKa = 11.84ASHH68 pKa = 6.07RR69 pKa = 11.84VGALITLFSLPSAGMQNHH87 pKa = 6.05IRR89 pKa = 11.84LADD92 pKa = 4.27RR93 pKa = 11.84SPEE96 pKa = 3.9AQIEE100 pKa = 4.18RR101 pKa = 11.84CEE103 pKa = 4.02IDD105 pKa = 3.34GFEE108 pKa = 4.47PGTYY112 pKa = 10.03RR113 pKa = 11.84LIPNARR119 pKa = 11.84ANLTANEE126 pKa = 3.72IAAYY130 pKa = 10.25ALLADD135 pKa = 5.13DD136 pKa = 6.14LPPTINNGTPYY147 pKa = 10.34VHH149 pKa = 7.43ADD151 pKa = 3.72VEE153 pKa = 4.47GQPCDD158 pKa = 3.97EE159 pKa = 4.8IEE161 pKa = 4.09QFLDD165 pKa = 2.71RR166 pKa = 11.84CYY168 pKa = 11.21SVLIQAWVMVCKK180 pKa = 10.67CMTAYY185 pKa = 9.79DD186 pKa = 4.06QPAGSADD193 pKa = 3.45RR194 pKa = 11.84RR195 pKa = 11.84FAKK198 pKa = 10.22YY199 pKa = 8.47QQQGRR204 pKa = 11.84LEE206 pKa = 4.21ARR208 pKa = 11.84YY209 pKa = 8.28MLQPEE214 pKa = 4.46AQRR217 pKa = 11.84LIQTAIRR224 pKa = 11.84KK225 pKa = 9.17SLVVRR230 pKa = 11.84QYY232 pKa = 10.98LTFEE236 pKa = 4.12LQLARR241 pKa = 11.84RR242 pKa = 11.84QGLLSNRR249 pKa = 11.84YY250 pKa = 8.0YY251 pKa = 11.82AMVGDD256 pKa = 3.41IGKK259 pKa = 10.26YY260 pKa = 9.45IEE262 pKa = 4.47NSGLTAFFLTLKK274 pKa = 10.27YY275 pKa = 10.86ALGTKK280 pKa = 9.03WSPLSLAAFTGEE292 pKa = 4.05LTKK295 pKa = 10.79LRR297 pKa = 11.84SLMMLYY303 pKa = 10.24RR304 pKa = 11.84DD305 pKa = 3.69LGEE308 pKa = 3.91QARR311 pKa = 11.84YY312 pKa = 8.75LALLEE317 pKa = 4.89APQIMDD323 pKa = 4.55FAPGGYY329 pKa = 9.29PLIFSYY335 pKa = 11.38AMGVGTVLDD344 pKa = 3.74VQMRR348 pKa = 11.84NYY350 pKa = 9.87TYY352 pKa = 11.25ARR354 pKa = 11.84PFLNGYY360 pKa = 7.57YY361 pKa = 9.8FQIGVEE367 pKa = 4.14TARR370 pKa = 11.84RR371 pKa = 11.84QQGTVDD377 pKa = 3.5NRR379 pKa = 11.84VADD382 pKa = 4.32DD383 pKa = 4.52LGLTPEE389 pKa = 3.96QRR391 pKa = 11.84TEE393 pKa = 3.78VTQLIDD399 pKa = 2.97RR400 pKa = 11.84LARR403 pKa = 11.84GRR405 pKa = 11.84GAGIPGGPVNPFVPPVQQQQPAAAYY430 pKa = 9.13EE431 pKa = 4.58DD432 pKa = 3.88IPALEE437 pKa = 4.64EE438 pKa = 4.19SDD440 pKa = 5.43DD441 pKa = 5.79DD442 pKa = 4.71GDD444 pKa = 3.87EE445 pKa = 4.51DD446 pKa = 5.2GGAGFQNGAQAPAARR461 pKa = 11.84QGGQNDD467 pKa = 3.7FRR469 pKa = 11.84VQPLQDD475 pKa = 4.84PIQAQLFMPLYY486 pKa = 8.72PQVSNIPNHH495 pKa = 5.8QNHH498 pKa = 6.27QINRR502 pKa = 11.84IGGMEE507 pKa = 4.03HH508 pKa = 7.16QDD510 pKa = 3.26LLRR513 pKa = 11.84YY514 pKa = 9.57NEE516 pKa = 4.83NGDD519 pKa = 3.68SQQDD523 pKa = 3.16ARR525 pKa = 11.84GEE527 pKa = 4.14HH528 pKa = 6.79GNTFPNNPNQNAQSQVGDD546 pKa = 3.37WDD548 pKa = 3.74EE549 pKa = 3.83
MM1 pKa = 7.49SSVLKK6 pKa = 10.65AFEE9 pKa = 4.31RR10 pKa = 11.84FTIEE14 pKa = 5.13QEE16 pKa = 3.95LQDD19 pKa = 3.9RR20 pKa = 11.84GEE22 pKa = 4.45EE23 pKa = 4.26GSIPPEE29 pKa = 4.03TLKK32 pKa = 11.08SAVKK36 pKa = 10.23VFVINTPNPTTRR48 pKa = 11.84YY49 pKa = 10.18HH50 pKa = 6.21MLNFCLRR57 pKa = 11.84IICSQNARR65 pKa = 11.84ASHH68 pKa = 6.07RR69 pKa = 11.84VGALITLFSLPSAGMQNHH87 pKa = 6.05IRR89 pKa = 11.84LADD92 pKa = 4.27RR93 pKa = 11.84SPEE96 pKa = 3.9AQIEE100 pKa = 4.18RR101 pKa = 11.84CEE103 pKa = 4.02IDD105 pKa = 3.34GFEE108 pKa = 4.47PGTYY112 pKa = 10.03RR113 pKa = 11.84LIPNARR119 pKa = 11.84ANLTANEE126 pKa = 3.72IAAYY130 pKa = 10.25ALLADD135 pKa = 5.13DD136 pKa = 6.14LPPTINNGTPYY147 pKa = 10.34VHH149 pKa = 7.43ADD151 pKa = 3.72VEE153 pKa = 4.47GQPCDD158 pKa = 3.97EE159 pKa = 4.8IEE161 pKa = 4.09QFLDD165 pKa = 2.71RR166 pKa = 11.84CYY168 pKa = 11.21SVLIQAWVMVCKK180 pKa = 10.67CMTAYY185 pKa = 9.79DD186 pKa = 4.06QPAGSADD193 pKa = 3.45RR194 pKa = 11.84RR195 pKa = 11.84FAKK198 pKa = 10.22YY199 pKa = 8.47QQQGRR204 pKa = 11.84LEE206 pKa = 4.21ARR208 pKa = 11.84YY209 pKa = 8.28MLQPEE214 pKa = 4.46AQRR217 pKa = 11.84LIQTAIRR224 pKa = 11.84KK225 pKa = 9.17SLVVRR230 pKa = 11.84QYY232 pKa = 10.98LTFEE236 pKa = 4.12LQLARR241 pKa = 11.84RR242 pKa = 11.84QGLLSNRR249 pKa = 11.84YY250 pKa = 8.0YY251 pKa = 11.82AMVGDD256 pKa = 3.41IGKK259 pKa = 10.26YY260 pKa = 9.45IEE262 pKa = 4.47NSGLTAFFLTLKK274 pKa = 10.27YY275 pKa = 10.86ALGTKK280 pKa = 9.03WSPLSLAAFTGEE292 pKa = 4.05LTKK295 pKa = 10.79LRR297 pKa = 11.84SLMMLYY303 pKa = 10.24RR304 pKa = 11.84DD305 pKa = 3.69LGEE308 pKa = 3.91QARR311 pKa = 11.84YY312 pKa = 8.75LALLEE317 pKa = 4.89APQIMDD323 pKa = 4.55FAPGGYY329 pKa = 9.29PLIFSYY335 pKa = 11.38AMGVGTVLDD344 pKa = 3.74VQMRR348 pKa = 11.84NYY350 pKa = 9.87TYY352 pKa = 11.25ARR354 pKa = 11.84PFLNGYY360 pKa = 7.57YY361 pKa = 9.8FQIGVEE367 pKa = 4.14TARR370 pKa = 11.84RR371 pKa = 11.84QQGTVDD377 pKa = 3.5NRR379 pKa = 11.84VADD382 pKa = 4.32DD383 pKa = 4.52LGLTPEE389 pKa = 3.96QRR391 pKa = 11.84TEE393 pKa = 3.78VTQLIDD399 pKa = 2.97RR400 pKa = 11.84LARR403 pKa = 11.84GRR405 pKa = 11.84GAGIPGGPVNPFVPPVQQQQPAAAYY430 pKa = 9.13EE431 pKa = 4.58DD432 pKa = 3.88IPALEE437 pKa = 4.64EE438 pKa = 4.19SDD440 pKa = 5.43DD441 pKa = 5.79DD442 pKa = 4.71GDD444 pKa = 3.87EE445 pKa = 4.51DD446 pKa = 5.2GGAGFQNGAQAPAARR461 pKa = 11.84QGGQNDD467 pKa = 3.7FRR469 pKa = 11.84VQPLQDD475 pKa = 4.84PIQAQLFMPLYY486 pKa = 8.72PQVSNIPNHH495 pKa = 5.8QNHH498 pKa = 6.27QINRR502 pKa = 11.84IGGMEE507 pKa = 4.03HH508 pKa = 7.16QDD510 pKa = 3.26LLRR513 pKa = 11.84YY514 pKa = 9.57NEE516 pKa = 4.83NGDD519 pKa = 3.68SQQDD523 pKa = 3.16ARR525 pKa = 11.84GEE527 pKa = 4.14HH528 pKa = 6.79GNTFPNNPNQNAQSQVGDD546 pKa = 3.37WDD548 pKa = 3.74EE549 pKa = 3.83
Molecular weight: 61.37 kDa
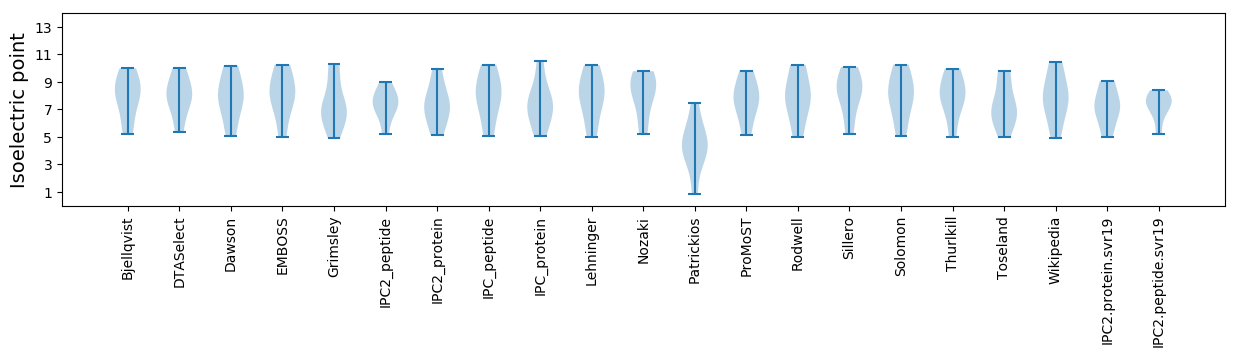
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P30928|V_MUMPM Non-structural protein V OS=Mumps virus (strain Miyahara vaccine) OX=11171 GN=P/V PE=1 SV=1
MM1 pKa = 7.73PAIQPPLYY9 pKa = 8.82PTFLLLILLSLIITLYY25 pKa = 9.9VWIISTITYY34 pKa = 6.96KK35 pKa = 10.16TAVRR39 pKa = 11.84HH40 pKa = 5.43AALHH44 pKa = 5.06QRR46 pKa = 11.84SFSRR50 pKa = 11.84WSLDD54 pKa = 2.96HH55 pKa = 6.98SLL57 pKa = 4.1
MM1 pKa = 7.73PAIQPPLYY9 pKa = 8.82PTFLLLILLSLIITLYY25 pKa = 9.9VWIISTITYY34 pKa = 6.96KK35 pKa = 10.16TAVRR39 pKa = 11.84HH40 pKa = 5.43AALHH44 pKa = 5.06QRR46 pKa = 11.84SFSRR50 pKa = 11.84WSLDD54 pKa = 2.96HH55 pKa = 6.98SLL57 pKa = 4.1
Molecular weight: 6.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4977 |
57 |
2261 |
622.1 |
69.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.631 ± 0.789 | 1.929 ± 0.292 |
4.561 ± 0.451 | 4.842 ± 0.571 |
3.355 ± 0.243 | 5.646 ± 1.046 |
1.869 ± 0.206 | 7.836 ± 0.78 |
4.26 ± 0.567 | 10.589 ± 1.373 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.311 ± 0.147 | 5.204 ± 0.532 |
5.365 ± 0.618 | 5.083 ± 0.815 |
5.083 ± 0.423 | 8.62 ± 0.792 |
6.409 ± 0.449 | 5.767 ± 0.353 |
1.125 ± 0.245 | 3.516 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
