
Halorubrum saccharovorum
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halorubrum
Average proteome isoelectric point is 4.91
Get precalculated fractions of proteins
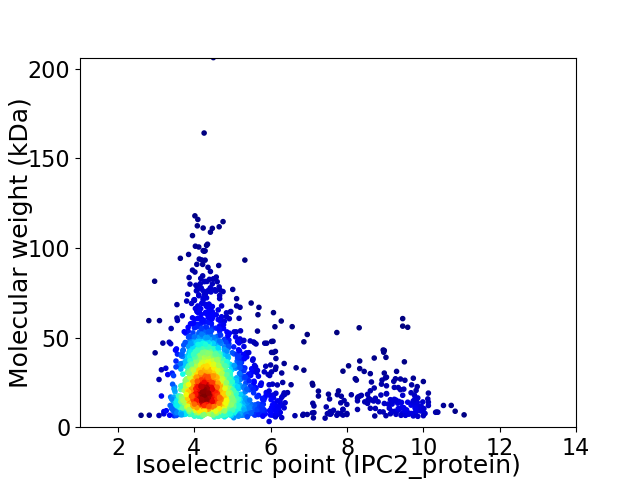
Virtual 2D-PAGE plot for 2334 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A081EX71|A0A081EX71_9EURY Hydrolase OS=Halorubrum saccharovorum OX=2248 GN=FK85_11285 PE=4 SV=1
MM1 pKa = 7.7EE2 pKa = 5.18SPRR5 pKa = 11.84KK6 pKa = 8.47LTTLIIIAGMVALAGCTGVIPGDD29 pKa = 3.74PAPADD34 pKa = 3.65PEE36 pKa = 4.22PANGSSTVTTNGSVEE51 pKa = 3.71VHH53 pKa = 6.43YY54 pKa = 10.97INVGQSVSTLITSPNGDD71 pKa = 3.28TMLVDD76 pKa = 3.67TGHH79 pKa = 6.22YY80 pKa = 10.12NDD82 pKa = 5.41DD83 pKa = 4.08GEE85 pKa = 4.98HH86 pKa = 5.36VLAYY90 pKa = 9.61LQRR93 pKa = 11.84HH94 pKa = 6.22DD95 pKa = 3.99ISRR98 pKa = 11.84IDD100 pKa = 3.62HH101 pKa = 6.62LVTSHH106 pKa = 6.88SDD108 pKa = 3.1ADD110 pKa = 4.29HH111 pKa = 6.8IGGNAAIIKK120 pKa = 9.64YY121 pKa = 10.25YY122 pKa = 8.54EE123 pKa = 4.14TEE125 pKa = 3.79ADD127 pKa = 4.24GIGAVYY133 pKa = 10.42DD134 pKa = 3.92PGIASSTQTYY144 pKa = 10.78AEE146 pKa = 4.13YY147 pKa = 10.98LDD149 pKa = 5.56AIEE152 pKa = 4.53AHH154 pKa = 7.09DD155 pKa = 3.69VTLYY159 pKa = 7.32EE160 pKa = 4.17TRR162 pKa = 11.84EE163 pKa = 3.69GDD165 pKa = 3.64TIDD168 pKa = 4.74FGDD171 pKa = 3.6VDD173 pKa = 4.51VDD175 pKa = 3.79VLGPPDD181 pKa = 4.8PYY183 pKa = 11.22LEE185 pKa = 4.36SEE187 pKa = 4.43DD188 pKa = 4.4RR189 pKa = 11.84NEE191 pKa = 3.74NSIVLKK197 pKa = 9.16LTHH200 pKa = 6.83GEE202 pKa = 4.16TSFLLSGDD210 pKa = 4.15AEE212 pKa = 4.88DD213 pKa = 4.69DD214 pKa = 3.69QEE216 pKa = 6.29AYY218 pKa = 10.68LVDD221 pKa = 4.11EE222 pKa = 4.89YY223 pKa = 11.57GSEE226 pKa = 4.27LEE228 pKa = 4.18STILKK233 pKa = 10.17AGHH236 pKa = 6.74HH237 pKa = 6.51GSASSSSGAFLDD249 pKa = 4.76AVDD252 pKa = 4.16PQAVVISSAYY262 pKa = 9.72DD263 pKa = 3.14SQYY266 pKa = 10.55GHH268 pKa = 6.39PHH270 pKa = 6.01EE271 pKa = 4.92EE272 pKa = 3.94VLEE275 pKa = 4.11RR276 pKa = 11.84LAEE279 pKa = 4.25RR280 pKa = 11.84SHH282 pKa = 6.11PAYY285 pKa = 7.58WTATHH290 pKa = 6.39GNIVLTSDD298 pKa = 3.46GQTVTVQTQRR308 pKa = 11.84DD309 pKa = 4.15SPTDD313 pKa = 3.56PSTLRR318 pKa = 11.84EE319 pKa = 4.07GDD321 pKa = 4.28PIDD324 pKa = 4.01VGVPDD329 pKa = 4.34AVSDD333 pKa = 3.76RR334 pKa = 11.84ARR336 pKa = 11.84IEE338 pKa = 4.11SDD340 pKa = 2.8GTTSIDD346 pKa = 3.26TGSNEE351 pKa = 4.27TPSDD355 pKa = 3.67ADD357 pKa = 3.59SEE359 pKa = 4.49TDD361 pKa = 3.15EE362 pKa = 4.36TAAEE366 pKa = 4.26AGLTVAEE373 pKa = 4.35IHH375 pKa = 6.93ADD377 pKa = 3.3AAGDD381 pKa = 4.69DD382 pKa = 3.82RR383 pKa = 11.84DD384 pKa = 4.07NLNDD388 pKa = 3.53EE389 pKa = 4.46YY390 pKa = 11.73VVFEE394 pKa = 4.04NTGNEE399 pKa = 4.09TLDD402 pKa = 3.49LSGWTIEE409 pKa = 4.66DD410 pKa = 3.47EE411 pKa = 4.14VGQRR415 pKa = 11.84YY416 pKa = 6.41TVPEE420 pKa = 4.38GFEE423 pKa = 4.05LAPGQTVTLHH433 pKa = 6.02TGSGTSTTTEE443 pKa = 4.65LYY445 pKa = 8.1WASGSPIWNNDD456 pKa = 2.31GDD458 pKa = 4.18IVIVSNANGEE468 pKa = 4.33RR469 pKa = 11.84VLEE472 pKa = 4.13EE473 pKa = 4.32TYY475 pKa = 11.25SS476 pKa = 3.34
MM1 pKa = 7.7EE2 pKa = 5.18SPRR5 pKa = 11.84KK6 pKa = 8.47LTTLIIIAGMVALAGCTGVIPGDD29 pKa = 3.74PAPADD34 pKa = 3.65PEE36 pKa = 4.22PANGSSTVTTNGSVEE51 pKa = 3.71VHH53 pKa = 6.43YY54 pKa = 10.97INVGQSVSTLITSPNGDD71 pKa = 3.28TMLVDD76 pKa = 3.67TGHH79 pKa = 6.22YY80 pKa = 10.12NDD82 pKa = 5.41DD83 pKa = 4.08GEE85 pKa = 4.98HH86 pKa = 5.36VLAYY90 pKa = 9.61LQRR93 pKa = 11.84HH94 pKa = 6.22DD95 pKa = 3.99ISRR98 pKa = 11.84IDD100 pKa = 3.62HH101 pKa = 6.62LVTSHH106 pKa = 6.88SDD108 pKa = 3.1ADD110 pKa = 4.29HH111 pKa = 6.8IGGNAAIIKK120 pKa = 9.64YY121 pKa = 10.25YY122 pKa = 8.54EE123 pKa = 4.14TEE125 pKa = 3.79ADD127 pKa = 4.24GIGAVYY133 pKa = 10.42DD134 pKa = 3.92PGIASSTQTYY144 pKa = 10.78AEE146 pKa = 4.13YY147 pKa = 10.98LDD149 pKa = 5.56AIEE152 pKa = 4.53AHH154 pKa = 7.09DD155 pKa = 3.69VTLYY159 pKa = 7.32EE160 pKa = 4.17TRR162 pKa = 11.84EE163 pKa = 3.69GDD165 pKa = 3.64TIDD168 pKa = 4.74FGDD171 pKa = 3.6VDD173 pKa = 4.51VDD175 pKa = 3.79VLGPPDD181 pKa = 4.8PYY183 pKa = 11.22LEE185 pKa = 4.36SEE187 pKa = 4.43DD188 pKa = 4.4RR189 pKa = 11.84NEE191 pKa = 3.74NSIVLKK197 pKa = 9.16LTHH200 pKa = 6.83GEE202 pKa = 4.16TSFLLSGDD210 pKa = 4.15AEE212 pKa = 4.88DD213 pKa = 4.69DD214 pKa = 3.69QEE216 pKa = 6.29AYY218 pKa = 10.68LVDD221 pKa = 4.11EE222 pKa = 4.89YY223 pKa = 11.57GSEE226 pKa = 4.27LEE228 pKa = 4.18STILKK233 pKa = 10.17AGHH236 pKa = 6.74HH237 pKa = 6.51GSASSSSGAFLDD249 pKa = 4.76AVDD252 pKa = 4.16PQAVVISSAYY262 pKa = 9.72DD263 pKa = 3.14SQYY266 pKa = 10.55GHH268 pKa = 6.39PHH270 pKa = 6.01EE271 pKa = 4.92EE272 pKa = 3.94VLEE275 pKa = 4.11RR276 pKa = 11.84LAEE279 pKa = 4.25RR280 pKa = 11.84SHH282 pKa = 6.11PAYY285 pKa = 7.58WTATHH290 pKa = 6.39GNIVLTSDD298 pKa = 3.46GQTVTVQTQRR308 pKa = 11.84DD309 pKa = 4.15SPTDD313 pKa = 3.56PSTLRR318 pKa = 11.84EE319 pKa = 4.07GDD321 pKa = 4.28PIDD324 pKa = 4.01VGVPDD329 pKa = 4.34AVSDD333 pKa = 3.76RR334 pKa = 11.84ARR336 pKa = 11.84IEE338 pKa = 4.11SDD340 pKa = 2.8GTTSIDD346 pKa = 3.26TGSNEE351 pKa = 4.27TPSDD355 pKa = 3.67ADD357 pKa = 3.59SEE359 pKa = 4.49TDD361 pKa = 3.15EE362 pKa = 4.36TAAEE366 pKa = 4.26AGLTVAEE373 pKa = 4.35IHH375 pKa = 6.93ADD377 pKa = 3.3AAGDD381 pKa = 4.69DD382 pKa = 3.82RR383 pKa = 11.84DD384 pKa = 4.07NLNDD388 pKa = 3.53EE389 pKa = 4.46YY390 pKa = 11.73VVFEE394 pKa = 4.04NTGNEE399 pKa = 4.09TLDD402 pKa = 3.49LSGWTIEE409 pKa = 4.66DD410 pKa = 3.47EE411 pKa = 4.14VGQRR415 pKa = 11.84YY416 pKa = 6.41TVPEE420 pKa = 4.38GFEE423 pKa = 4.05LAPGQTVTLHH433 pKa = 6.02TGSGTSTTTEE443 pKa = 4.65LYY445 pKa = 8.1WASGSPIWNNDD456 pKa = 2.31GDD458 pKa = 4.18IVIVSNANGEE468 pKa = 4.33RR469 pKa = 11.84VLEE472 pKa = 4.13EE473 pKa = 4.32TYY475 pKa = 11.25SS476 pKa = 3.34
Molecular weight: 50.7 kDa
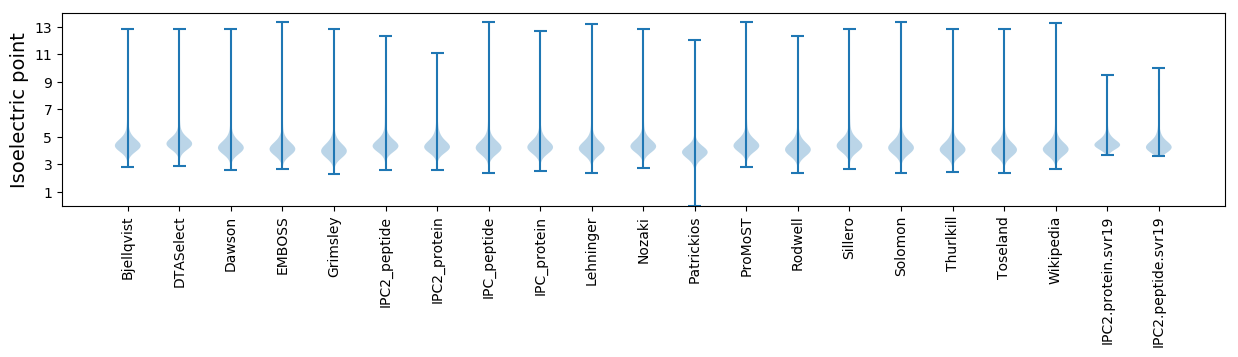
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F8BHS3|A0A0F8BHS3_9EURY NUDIX hydrolase OS=Halorubrum saccharovorum OX=2248 GN=FK85_25910 PE=4 SV=1
RR1 pKa = 7.3GRR3 pKa = 11.84HH4 pKa = 4.35QAGASATAPATARR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84GAGIARR26 pKa = 11.84RR27 pKa = 11.84APRR30 pKa = 11.84DD31 pKa = 2.9RR32 pKa = 11.84RR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84VRR41 pKa = 11.84ASGRR45 pKa = 11.84PSPARR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84PGRR56 pKa = 11.84PRR58 pKa = 11.84EE59 pKa = 3.87RR60 pKa = 11.84RR61 pKa = 3.39
RR1 pKa = 7.3GRR3 pKa = 11.84HH4 pKa = 4.35QAGASATAPATARR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84GAGIARR26 pKa = 11.84RR27 pKa = 11.84APRR30 pKa = 11.84DD31 pKa = 2.9RR32 pKa = 11.84RR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84VRR41 pKa = 11.84ASGRR45 pKa = 11.84PSPARR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84PGRR56 pKa = 11.84PRR58 pKa = 11.84EE59 pKa = 3.87RR60 pKa = 11.84RR61 pKa = 3.39
Molecular weight: 6.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
584998 |
29 |
1869 |
250.6 |
27.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.04 ± 0.081 | 0.66 ± 0.017 |
8.879 ± 0.067 | 8.719 ± 0.076 |
3.323 ± 0.034 | 8.811 ± 0.058 |
1.908 ± 0.027 | 4.151 ± 0.038 |
1.855 ± 0.037 | 8.641 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.745 ± 0.025 | 2.318 ± 0.031 |
4.628 ± 0.033 | 2.245 ± 0.035 |
6.68 ± 0.048 | 5.538 ± 0.039 |
6.118 ± 0.038 | 9.063 ± 0.057 |
1.083 ± 0.017 | 2.595 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
