
Crocuta crocuta papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Lambdapapillomavirus; Lambdapapillomavirus 5
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
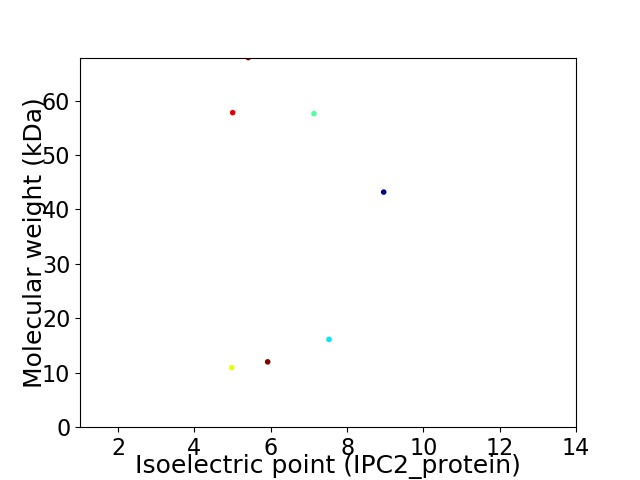
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7EZS5|J7EZS5_9PAPI Major capsid protein L1 OS=Crocuta crocuta papillomavirus 1 OX=1104917 GN=L1 PE=3 SV=1
MM1 pKa = 7.48RR2 pKa = 11.84GEE4 pKa = 4.3RR5 pKa = 11.84PTLQEE10 pKa = 3.59IVLTEE15 pKa = 4.12LPEE18 pKa = 5.85HH19 pKa = 7.04IDD21 pKa = 4.05LEE23 pKa = 4.66CHH25 pKa = 4.61EE26 pKa = 4.4QMPEE30 pKa = 3.87EE31 pKa = 4.29EE32 pKa = 4.68EE33 pKa = 4.09EE34 pKa = 4.22EE35 pKa = 4.27EE36 pKa = 4.94PISRR40 pKa = 11.84DD41 pKa = 3.27CFRR44 pKa = 11.84VATCCGYY51 pKa = 10.16CRR53 pKa = 11.84KK54 pKa = 9.42EE55 pKa = 3.93LRR57 pKa = 11.84FLCLSSAAQVRR68 pKa = 11.84SLEE71 pKa = 4.12SLLLQDD77 pKa = 5.18FSFICTSCVVKK88 pKa = 10.75KK89 pKa = 10.51KK90 pKa = 10.4LNHH93 pKa = 6.32GGG95 pKa = 3.38
MM1 pKa = 7.48RR2 pKa = 11.84GEE4 pKa = 4.3RR5 pKa = 11.84PTLQEE10 pKa = 3.59IVLTEE15 pKa = 4.12LPEE18 pKa = 5.85HH19 pKa = 7.04IDD21 pKa = 4.05LEE23 pKa = 4.66CHH25 pKa = 4.61EE26 pKa = 4.4QMPEE30 pKa = 3.87EE31 pKa = 4.29EE32 pKa = 4.68EE33 pKa = 4.09EE34 pKa = 4.22EE35 pKa = 4.27EE36 pKa = 4.94PISRR40 pKa = 11.84DD41 pKa = 3.27CFRR44 pKa = 11.84VATCCGYY51 pKa = 10.16CRR53 pKa = 11.84KK54 pKa = 9.42EE55 pKa = 3.93LRR57 pKa = 11.84FLCLSSAAQVRR68 pKa = 11.84SLEE71 pKa = 4.12SLLLQDD77 pKa = 5.18FSFICTSCVVKK88 pKa = 10.75KK89 pKa = 10.51KK90 pKa = 10.4LNHH93 pKa = 6.32GGG95 pKa = 3.38
Molecular weight: 10.93 kDa
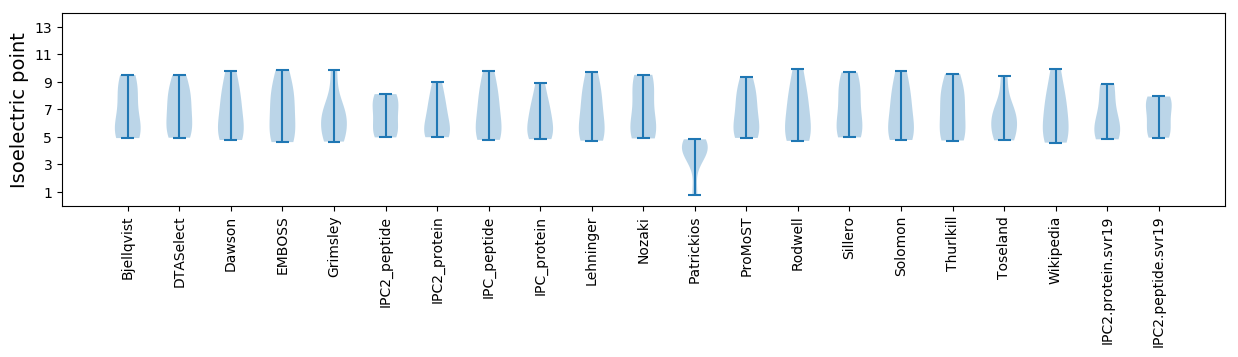
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7F282|J7F282_9PAPI Protein E6 OS=Crocuta crocuta papillomavirus 1 OX=1104917 GN=E6 PE=3 SV=1
MM1 pKa = 7.12EE2 pKa = 4.97TLSQLLEE9 pKa = 4.13SLQNDD14 pKa = 3.62MLTLYY19 pKa = 10.51EE20 pKa = 4.64KK21 pKa = 10.98DD22 pKa = 3.32SDD24 pKa = 4.03RR25 pKa = 11.84LTDD28 pKa = 5.5QITHH32 pKa = 4.64WHH34 pKa = 6.65LNRR37 pKa = 11.84KK38 pKa = 7.42EE39 pKa = 3.61QTLYY43 pKa = 10.55YY44 pKa = 10.2GARR47 pKa = 11.84QRR49 pKa = 11.84GITRR53 pKa = 11.84IGYY56 pKa = 8.13NHH58 pKa = 6.12VPSLAASQQRR68 pKa = 11.84AKK70 pKa = 10.5QAIEE74 pKa = 3.67QEE76 pKa = 4.49LILRR80 pKa = 11.84TLLDD84 pKa = 3.65SEE86 pKa = 4.84FGEE89 pKa = 4.61EE90 pKa = 4.28PWNLMQTSRR99 pKa = 11.84EE100 pKa = 3.89RR101 pKa = 11.84LLAPPAYY108 pKa = 9.23CFKK111 pKa = 10.91KK112 pKa = 10.23GGHH115 pKa = 6.32NIDD118 pKa = 3.28VKK120 pKa = 11.13FNNDD124 pKa = 2.83SQNIARR130 pKa = 11.84YY131 pKa = 8.55VLWDD135 pKa = 4.47FIYY138 pKa = 11.14YY139 pKa = 9.63QDD141 pKa = 5.73SEE143 pKa = 4.77DD144 pKa = 3.04KK145 pKa = 9.6WRR147 pKa = 11.84VSKK150 pKa = 11.04GYY152 pKa = 10.45VDD154 pKa = 3.56EE155 pKa = 4.65TGLYY159 pKa = 10.09YY160 pKa = 9.91EE161 pKa = 4.71TEE163 pKa = 4.29LGIKK167 pKa = 9.32IYY169 pKa = 10.62YY170 pKa = 10.12VNFHH174 pKa = 7.38DD175 pKa = 5.04EE176 pKa = 3.7AAKK179 pKa = 10.69YY180 pKa = 10.31GNSDD184 pKa = 3.02VYY186 pKa = 11.04EE187 pKa = 4.26VVKK190 pKa = 10.14TPLTTPVSTSTSGLNTASPGPHH212 pKa = 6.56SDD214 pKa = 2.59SGTPRR219 pKa = 11.84RR220 pKa = 11.84KK221 pKa = 8.24TSTPVKK227 pKa = 9.95SRR229 pKa = 11.84KK230 pKa = 9.26KK231 pKa = 9.82SGRR234 pKa = 11.84HH235 pKa = 5.54ALPKK239 pKa = 9.37TPVGRR244 pKa = 11.84RR245 pKa = 11.84FGRR248 pKa = 11.84RR249 pKa = 11.84KK250 pKa = 10.09GEE252 pKa = 4.09RR253 pKa = 11.84VSTATPSSKK262 pKa = 10.3PPSPEE267 pKa = 3.78QIGSSHH273 pKa = 5.86QSVPVRR279 pKa = 11.84GGGGRR284 pKa = 11.84LGRR287 pKa = 11.84LLAEE291 pKa = 4.8ARR293 pKa = 11.84DD294 pKa = 3.98PPCIVIRR301 pKa = 11.84GDD303 pKa = 3.65PNTVKK308 pKa = 10.68CLRR311 pKa = 11.84YY312 pKa = 9.59RR313 pKa = 11.84FKK315 pKa = 10.78QQYY318 pKa = 9.82QNLFWGVSTTWSWTWKK334 pKa = 10.29GSEE337 pKa = 3.66QSKK340 pKa = 9.41RR341 pKa = 11.84SRR343 pKa = 11.84IIFLFTDD350 pKa = 3.48DD351 pKa = 4.01DD352 pKa = 3.83QRR354 pKa = 11.84EE355 pKa = 4.23KK356 pKa = 11.02FLNTVKK362 pKa = 10.53LPKK365 pKa = 9.94SVSYY369 pKa = 10.2FLGSFNDD376 pKa = 3.93YY377 pKa = 11.27
MM1 pKa = 7.12EE2 pKa = 4.97TLSQLLEE9 pKa = 4.13SLQNDD14 pKa = 3.62MLTLYY19 pKa = 10.51EE20 pKa = 4.64KK21 pKa = 10.98DD22 pKa = 3.32SDD24 pKa = 4.03RR25 pKa = 11.84LTDD28 pKa = 5.5QITHH32 pKa = 4.64WHH34 pKa = 6.65LNRR37 pKa = 11.84KK38 pKa = 7.42EE39 pKa = 3.61QTLYY43 pKa = 10.55YY44 pKa = 10.2GARR47 pKa = 11.84QRR49 pKa = 11.84GITRR53 pKa = 11.84IGYY56 pKa = 8.13NHH58 pKa = 6.12VPSLAASQQRR68 pKa = 11.84AKK70 pKa = 10.5QAIEE74 pKa = 3.67QEE76 pKa = 4.49LILRR80 pKa = 11.84TLLDD84 pKa = 3.65SEE86 pKa = 4.84FGEE89 pKa = 4.61EE90 pKa = 4.28PWNLMQTSRR99 pKa = 11.84EE100 pKa = 3.89RR101 pKa = 11.84LLAPPAYY108 pKa = 9.23CFKK111 pKa = 10.91KK112 pKa = 10.23GGHH115 pKa = 6.32NIDD118 pKa = 3.28VKK120 pKa = 11.13FNNDD124 pKa = 2.83SQNIARR130 pKa = 11.84YY131 pKa = 8.55VLWDD135 pKa = 4.47FIYY138 pKa = 11.14YY139 pKa = 9.63QDD141 pKa = 5.73SEE143 pKa = 4.77DD144 pKa = 3.04KK145 pKa = 9.6WRR147 pKa = 11.84VSKK150 pKa = 11.04GYY152 pKa = 10.45VDD154 pKa = 3.56EE155 pKa = 4.65TGLYY159 pKa = 10.09YY160 pKa = 9.91EE161 pKa = 4.71TEE163 pKa = 4.29LGIKK167 pKa = 9.32IYY169 pKa = 10.62YY170 pKa = 10.12VNFHH174 pKa = 7.38DD175 pKa = 5.04EE176 pKa = 3.7AAKK179 pKa = 10.69YY180 pKa = 10.31GNSDD184 pKa = 3.02VYY186 pKa = 11.04EE187 pKa = 4.26VVKK190 pKa = 10.14TPLTTPVSTSTSGLNTASPGPHH212 pKa = 6.56SDD214 pKa = 2.59SGTPRR219 pKa = 11.84RR220 pKa = 11.84KK221 pKa = 8.24TSTPVKK227 pKa = 9.95SRR229 pKa = 11.84KK230 pKa = 9.26KK231 pKa = 9.82SGRR234 pKa = 11.84HH235 pKa = 5.54ALPKK239 pKa = 9.37TPVGRR244 pKa = 11.84RR245 pKa = 11.84FGRR248 pKa = 11.84RR249 pKa = 11.84KK250 pKa = 10.09GEE252 pKa = 4.09RR253 pKa = 11.84VSTATPSSKK262 pKa = 10.3PPSPEE267 pKa = 3.78QIGSSHH273 pKa = 5.86QSVPVRR279 pKa = 11.84GGGGRR284 pKa = 11.84LGRR287 pKa = 11.84LLAEE291 pKa = 4.8ARR293 pKa = 11.84DD294 pKa = 3.98PPCIVIRR301 pKa = 11.84GDD303 pKa = 3.65PNTVKK308 pKa = 10.68CLRR311 pKa = 11.84YY312 pKa = 9.59RR313 pKa = 11.84FKK315 pKa = 10.78QQYY318 pKa = 9.82QNLFWGVSTTWSWTWKK334 pKa = 10.29GSEE337 pKa = 3.66QSKK340 pKa = 9.41RR341 pKa = 11.84SRR343 pKa = 11.84IIFLFTDD350 pKa = 3.48DD351 pKa = 4.01DD352 pKa = 3.83QRR354 pKa = 11.84EE355 pKa = 4.23KK356 pKa = 11.02FLNTVKK362 pKa = 10.53LPKK365 pKa = 9.94SVSYY369 pKa = 10.2FLGSFNDD376 pKa = 3.93YY377 pKa = 11.27
Molecular weight: 43.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2351 |
95 |
590 |
335.9 |
37.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.572 ± 0.593 | 2.467 ± 0.94 |
5.87 ± 0.395 | 6.423 ± 0.789 |
4.764 ± 0.352 | 5.827 ± 0.716 |
2.212 ± 0.267 | 5.359 ± 0.614 |
6.21 ± 0.561 | 9.485 ± 1.23 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.276 ± 0.231 | 4.679 ± 0.577 |
5.955 ± 0.819 | 3.913 ± 0.414 |
5.317 ± 0.616 | 7.571 ± 0.735 |
6.423 ± 0.515 | 5.827 ± 0.59 |
1.446 ± 0.347 | 3.403 ± 0.477 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
