
human papillomavirus 108
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 6
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|C0MHS1|C0MHS1_9PAPI Replication protein E1 OS=human papillomavirus 108 OX=565537 GN=E1 PE=3 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84GKK4 pKa = 10.55APTIKK9 pKa = 10.73DD10 pKa = 3.02VDD12 pKa = 4.41LEE14 pKa = 4.28LHH16 pKa = 6.52SLILPEE22 pKa = 4.48NLLCGEE28 pKa = 4.15EE29 pKa = 4.14VTFEE33 pKa = 4.19KK34 pKa = 10.81EE35 pKa = 3.84EE36 pKa = 3.93QEE38 pKa = 4.36EE39 pKa = 4.07QDD41 pKa = 3.19KK42 pKa = 11.23CPYY45 pKa = 10.1RR46 pKa = 11.84VVTEE50 pKa = 4.52CGVCHH55 pKa = 7.04KK56 pKa = 9.63GLCMYY61 pKa = 10.68VLATDD66 pKa = 3.37EE67 pKa = 4.8GIRR70 pKa = 11.84GFEE73 pKa = 3.83QQLCGDD79 pKa = 4.84LDD81 pKa = 4.05IVCPACAKK89 pKa = 9.58QRR91 pKa = 11.84EE92 pKa = 4.5RR93 pKa = 11.84NGGQRR98 pKa = 11.84HH99 pKa = 4.79
MM1 pKa = 7.71RR2 pKa = 11.84GKK4 pKa = 10.55APTIKK9 pKa = 10.73DD10 pKa = 3.02VDD12 pKa = 4.41LEE14 pKa = 4.28LHH16 pKa = 6.52SLILPEE22 pKa = 4.48NLLCGEE28 pKa = 4.15EE29 pKa = 4.14VTFEE33 pKa = 4.19KK34 pKa = 10.81EE35 pKa = 3.84EE36 pKa = 3.93QEE38 pKa = 4.36EE39 pKa = 4.07QDD41 pKa = 3.19KK42 pKa = 11.23CPYY45 pKa = 10.1RR46 pKa = 11.84VVTEE50 pKa = 4.52CGVCHH55 pKa = 7.04KK56 pKa = 9.63GLCMYY61 pKa = 10.68VLATDD66 pKa = 3.37EE67 pKa = 4.8GIRR70 pKa = 11.84GFEE73 pKa = 3.83QQLCGDD79 pKa = 4.84LDD81 pKa = 4.05IVCPACAKK89 pKa = 9.58QRR91 pKa = 11.84EE92 pKa = 4.5RR93 pKa = 11.84NGGQRR98 pKa = 11.84HH99 pKa = 4.79
Molecular weight: 11.17 kDa
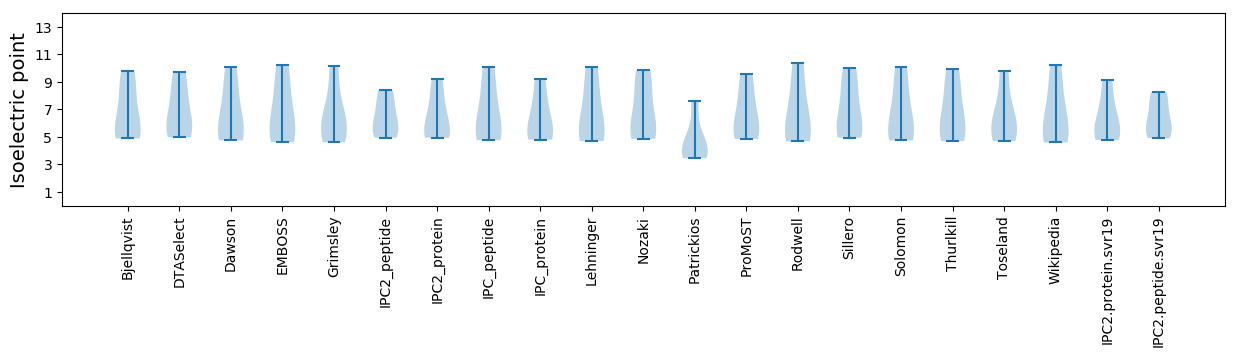
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0MHS3|C0MHS3_9PAPI Minor capsid protein L2 OS=human papillomavirus 108 OX=565537 GN=L2 PE=3 SV=1
MM1 pKa = 6.86EE2 pKa = 4.57TLVQRR7 pKa = 11.84FDD9 pKa = 3.64VVQDD13 pKa = 3.63KK14 pKa = 11.27LLGLYY19 pKa = 8.82EE20 pKa = 4.87AGHH23 pKa = 6.8TDD25 pKa = 4.16LDD27 pKa = 3.97SQIQYY32 pKa = 9.58WDD34 pKa = 3.5LVRR37 pKa = 11.84QEE39 pKa = 4.35NVLLNFARR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 5.71GIQMLGLQRR58 pKa = 11.84VPPLAISEE66 pKa = 4.6SKK68 pKa = 10.95AKK70 pKa = 8.47TAIMMRR76 pKa = 11.84LVLDD80 pKa = 3.97SLKK83 pKa = 10.56KK84 pKa = 10.68SPFGQEE90 pKa = 3.37PWGLTDD96 pKa = 3.83TSHH99 pKa = 6.83EE100 pKa = 4.71LYY102 pKa = 7.47TTPPEE107 pKa = 3.97NTFKK111 pKa = 11.02KK112 pKa = 10.35GGAQVEE118 pKa = 4.25VLYY121 pKa = 11.13DD122 pKa = 3.63NEE124 pKa = 4.24EE125 pKa = 4.15NKK127 pKa = 10.7AMPYY131 pKa = 9.3TKK133 pKa = 9.36WKK135 pKa = 9.49YY136 pKa = 10.31IYY138 pKa = 10.16YY139 pKa = 9.77QDD141 pKa = 5.95SEE143 pKa = 4.8DD144 pKa = 3.88LWHH147 pKa = 6.49KK148 pKa = 8.6THH150 pKa = 6.63AQVDD154 pKa = 3.83AKK156 pKa = 10.89GLYY159 pKa = 9.94FVDD162 pKa = 3.87TQMHH166 pKa = 5.38KK167 pKa = 10.14HH168 pKa = 5.99YY169 pKa = 10.49YY170 pKa = 8.43VQFDD174 pKa = 4.22KK175 pKa = 11.4EE176 pKa = 4.09AMQYY180 pKa = 7.95GHH182 pKa = 6.89KK183 pKa = 10.55GVWQVRR189 pKa = 11.84TGTDD193 pKa = 3.46TIFGASSVASTSRR206 pKa = 11.84GPDD209 pKa = 3.02ATSTDD214 pKa = 3.75PQAGRR219 pKa = 11.84LPEE222 pKa = 4.09RR223 pKa = 11.84KK224 pKa = 8.37RR225 pKa = 11.84KK226 pKa = 9.14RR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84YY231 pKa = 9.49PSTTTTATSEE241 pKa = 4.16SDD243 pKa = 3.67TEE245 pKa = 4.44EE246 pKa = 3.98STSDD250 pKa = 2.66NRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84KK255 pKa = 9.46RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84LRR260 pKa = 11.84EE261 pKa = 3.59RR262 pKa = 11.84KK263 pKa = 9.11HH264 pKa = 6.39RR265 pKa = 11.84SRR267 pKa = 11.84HH268 pKa = 3.75RR269 pKa = 11.84WGTGTEE275 pKa = 4.31SASDD279 pKa = 3.43TSASVGSRR287 pKa = 11.84HH288 pKa = 6.32RR289 pKa = 11.84SPNRR293 pKa = 11.84HH294 pKa = 5.19YY295 pKa = 10.44PSRR298 pKa = 11.84LARR301 pKa = 11.84LQAEE305 pKa = 4.34ARR307 pKa = 11.84DD308 pKa = 3.91PPILIVKK315 pKa = 9.76GPSNTLKK322 pKa = 10.24CWRR325 pKa = 11.84YY326 pKa = 8.85RR327 pKa = 11.84CKK329 pKa = 10.14TKK331 pKa = 11.07YY332 pKa = 10.1SGLYY336 pKa = 9.88VNISTGFTWVTGAGTEE352 pKa = 3.94GHH354 pKa = 6.51ARR356 pKa = 11.84ILVAFKK362 pKa = 11.2DD363 pKa = 3.41KK364 pKa = 9.94MQRR367 pKa = 11.84DD368 pKa = 4.0LFMQTVKK375 pKa = 10.66LPKK378 pKa = 9.53GTSCALGNLEE388 pKa = 4.65SLL390 pKa = 4.17
MM1 pKa = 6.86EE2 pKa = 4.57TLVQRR7 pKa = 11.84FDD9 pKa = 3.64VVQDD13 pKa = 3.63KK14 pKa = 11.27LLGLYY19 pKa = 8.82EE20 pKa = 4.87AGHH23 pKa = 6.8TDD25 pKa = 4.16LDD27 pKa = 3.97SQIQYY32 pKa = 9.58WDD34 pKa = 3.5LVRR37 pKa = 11.84QEE39 pKa = 4.35NVLLNFARR47 pKa = 11.84RR48 pKa = 11.84HH49 pKa = 5.71GIQMLGLQRR58 pKa = 11.84VPPLAISEE66 pKa = 4.6SKK68 pKa = 10.95AKK70 pKa = 8.47TAIMMRR76 pKa = 11.84LVLDD80 pKa = 3.97SLKK83 pKa = 10.56KK84 pKa = 10.68SPFGQEE90 pKa = 3.37PWGLTDD96 pKa = 3.83TSHH99 pKa = 6.83EE100 pKa = 4.71LYY102 pKa = 7.47TTPPEE107 pKa = 3.97NTFKK111 pKa = 11.02KK112 pKa = 10.35GGAQVEE118 pKa = 4.25VLYY121 pKa = 11.13DD122 pKa = 3.63NEE124 pKa = 4.24EE125 pKa = 4.15NKK127 pKa = 10.7AMPYY131 pKa = 9.3TKK133 pKa = 9.36WKK135 pKa = 9.49YY136 pKa = 10.31IYY138 pKa = 10.16YY139 pKa = 9.77QDD141 pKa = 5.95SEE143 pKa = 4.8DD144 pKa = 3.88LWHH147 pKa = 6.49KK148 pKa = 8.6THH150 pKa = 6.63AQVDD154 pKa = 3.83AKK156 pKa = 10.89GLYY159 pKa = 9.94FVDD162 pKa = 3.87TQMHH166 pKa = 5.38KK167 pKa = 10.14HH168 pKa = 5.99YY169 pKa = 10.49YY170 pKa = 8.43VQFDD174 pKa = 4.22KK175 pKa = 11.4EE176 pKa = 4.09AMQYY180 pKa = 7.95GHH182 pKa = 6.89KK183 pKa = 10.55GVWQVRR189 pKa = 11.84TGTDD193 pKa = 3.46TIFGASSVASTSRR206 pKa = 11.84GPDD209 pKa = 3.02ATSTDD214 pKa = 3.75PQAGRR219 pKa = 11.84LPEE222 pKa = 4.09RR223 pKa = 11.84KK224 pKa = 8.37RR225 pKa = 11.84KK226 pKa = 9.14RR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84YY231 pKa = 9.49PSTTTTATSEE241 pKa = 4.16SDD243 pKa = 3.67TEE245 pKa = 4.44EE246 pKa = 3.98STSDD250 pKa = 2.66NRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84KK255 pKa = 9.46RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84LRR260 pKa = 11.84EE261 pKa = 3.59RR262 pKa = 11.84KK263 pKa = 9.11HH264 pKa = 6.39RR265 pKa = 11.84SRR267 pKa = 11.84HH268 pKa = 3.75RR269 pKa = 11.84WGTGTEE275 pKa = 4.31SASDD279 pKa = 3.43TSASVGSRR287 pKa = 11.84HH288 pKa = 6.32RR289 pKa = 11.84SPNRR293 pKa = 11.84HH294 pKa = 5.19YY295 pKa = 10.44PSRR298 pKa = 11.84LARR301 pKa = 11.84LQAEE305 pKa = 4.34ARR307 pKa = 11.84DD308 pKa = 3.91PPILIVKK315 pKa = 9.76GPSNTLKK322 pKa = 10.24CWRR325 pKa = 11.84YY326 pKa = 8.85RR327 pKa = 11.84CKK329 pKa = 10.14TKK331 pKa = 11.07YY332 pKa = 10.1SGLYY336 pKa = 9.88VNISTGFTWVTGAGTEE352 pKa = 3.94GHH354 pKa = 6.51ARR356 pKa = 11.84ILVAFKK362 pKa = 11.2DD363 pKa = 3.41KK364 pKa = 9.94MQRR367 pKa = 11.84DD368 pKa = 4.0LFMQTVKK375 pKa = 10.66LPKK378 pKa = 9.53GTSCALGNLEE388 pKa = 4.65SLL390 pKa = 4.17
Molecular weight: 44.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2145 |
99 |
626 |
429.0 |
47.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.967 ± 0.401 | 2.005 ± 0.808 |
5.874 ± 0.198 | 5.781 ± 1.015 |
3.823 ± 0.395 | 6.34 ± 0.561 |
2.238 ± 0.26 | 4.709 ± 0.707 |
5.175 ± 0.932 | 8.065 ± 0.786 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.678 ± 0.41 | 3.776 ± 0.669 |
6.76 ± 1.175 | 4.755 ± 0.117 |
6.014 ± 1.017 | 8.298 ± 0.658 |
7.04 ± 0.706 | 7.086 ± 0.682 |
1.399 ± 0.317 | 3.217 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
