
Beihai hepe-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
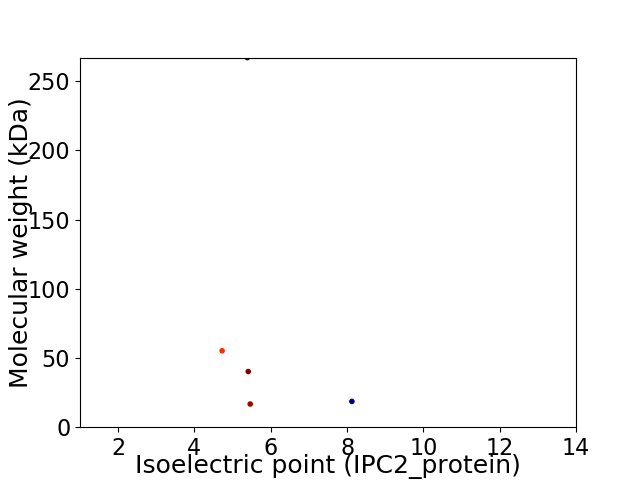
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
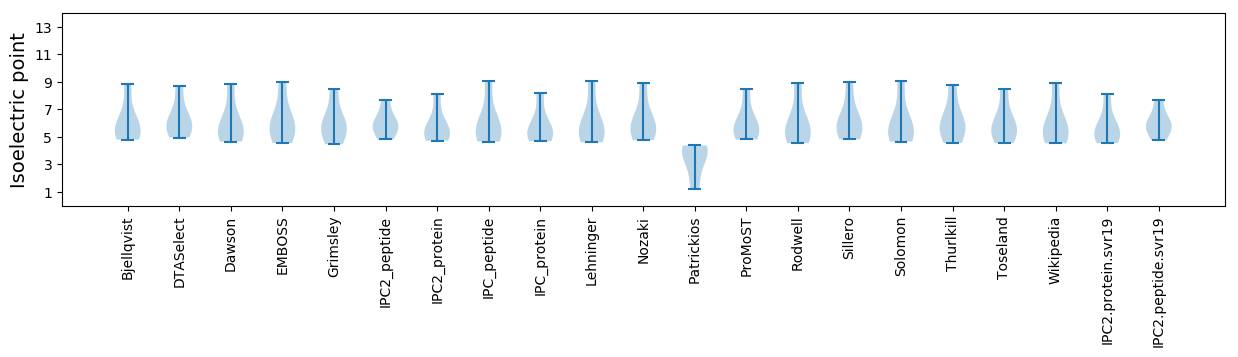
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJQ5|A0A1L3KJQ5_9VIRU Putative structural protein OS=Beihai hepe-like virus 10 OX=1922381 PE=4 SV=1
MM1 pKa = 7.15YY2 pKa = 10.56LILFYY7 pKa = 11.2AFFTLSSAQQLVEE20 pKa = 3.87LADD23 pKa = 3.39WQFNQIIARR32 pKa = 11.84EE33 pKa = 4.04NEE35 pKa = 3.6PNARR39 pKa = 11.84NFTCSDD45 pKa = 3.69FPNLVNWKK53 pKa = 10.14CGDD56 pKa = 3.73EE57 pKa = 4.45SPGRR61 pKa = 11.84AVARR65 pKa = 11.84VLPVGTVIPTFVPGPDD81 pKa = 3.25RR82 pKa = 11.84NSDD85 pKa = 3.26KK86 pKa = 11.13LGYY89 pKa = 8.45CTGSTTMGPYY99 pKa = 10.16NLSTIGKK106 pKa = 8.71FALEE110 pKa = 4.01LAQLEE115 pKa = 4.58VHH117 pKa = 6.51FNSHH121 pKa = 7.46FILSFDD127 pKa = 3.97IPPDD131 pKa = 3.41FDD133 pKa = 4.89FYY135 pKa = 11.74NDD137 pKa = 3.67PPYY140 pKa = 10.91LQAHH144 pKa = 5.1YY145 pKa = 9.25TGKK148 pKa = 9.66PLKK151 pKa = 9.07TQVSLKK157 pKa = 10.5QVDD160 pKa = 3.62NMLFALSQLSFEE172 pKa = 4.41QTSFDD177 pKa = 4.75DD178 pKa = 4.5KK179 pKa = 10.78EE180 pKa = 3.9QTAIDD185 pKa = 3.92VSALGMATGSVAYY198 pKa = 9.98DD199 pKa = 3.01QTTNVSSIFKK209 pKa = 8.82WFQYY213 pKa = 8.63EE214 pKa = 4.41FKK216 pKa = 11.02APNDD220 pKa = 3.7KK221 pKa = 10.15IKK223 pKa = 10.71RR224 pKa = 11.84IISPDD229 pKa = 2.51IYY231 pKa = 9.82YY232 pKa = 10.82TMFFKK237 pKa = 10.51DD238 pKa = 3.77DD239 pKa = 3.76KK240 pKa = 11.58ADD242 pKa = 3.74GGVSYY247 pKa = 9.15ITLPNPGLQLFADD260 pKa = 3.51AVEE263 pKa = 4.27RR264 pKa = 11.84TEE266 pKa = 4.49VALNYY271 pKa = 8.79QLRR274 pKa = 11.84SGITVSHH281 pKa = 6.87IDD283 pKa = 3.18IFSNIPQASYY293 pKa = 11.29NSPLTPLFSFFHH305 pKa = 6.04SDD307 pKa = 3.71KK308 pKa = 11.01YY309 pKa = 11.07ILSSWLRR316 pKa = 11.84FQNYY320 pKa = 5.74YY321 pKa = 8.73TYY323 pKa = 10.86KK324 pKa = 10.27GRR326 pKa = 11.84NITAQEE332 pKa = 3.99LNYY335 pKa = 10.78GITEE339 pKa = 4.04DD340 pKa = 3.82MRR342 pKa = 11.84NLVYY346 pKa = 9.71VQCHH350 pKa = 5.68EE351 pKa = 4.12RR352 pKa = 11.84PEE354 pKa = 4.02MCKK357 pKa = 10.58CEE359 pKa = 4.18GNYY362 pKa = 10.24SSQALAIGAFGVDD375 pKa = 3.09STYY378 pKa = 11.25TKK380 pKa = 10.8VPNTAIYY387 pKa = 9.93EE388 pKa = 4.16NQTFQVHH395 pKa = 5.44SPILHH400 pKa = 6.2HH401 pKa = 6.6VGSVPLEE408 pKa = 4.07GPYY411 pKa = 10.3TLFSSALPDD420 pKa = 3.78LPDD423 pKa = 3.38IKK425 pKa = 10.88ASNLPAWLPEE435 pKa = 4.24SVGEE439 pKa = 4.14EE440 pKa = 3.78IEE442 pKa = 4.42GVLNAVDD449 pKa = 3.92NFYY452 pKa = 11.47KK453 pKa = 10.31HH454 pKa = 6.75VKK456 pKa = 10.16DD457 pKa = 4.12FWEE460 pKa = 4.1ILQHH464 pKa = 7.25IEE466 pKa = 4.64EE467 pKa = 4.37IVEE470 pKa = 4.1VLKK473 pKa = 11.03DD474 pKa = 3.45IASLNPFGGSTPDD487 pKa = 3.27AVPVV491 pKa = 3.51
MM1 pKa = 7.15YY2 pKa = 10.56LILFYY7 pKa = 11.2AFFTLSSAQQLVEE20 pKa = 3.87LADD23 pKa = 3.39WQFNQIIARR32 pKa = 11.84EE33 pKa = 4.04NEE35 pKa = 3.6PNARR39 pKa = 11.84NFTCSDD45 pKa = 3.69FPNLVNWKK53 pKa = 10.14CGDD56 pKa = 3.73EE57 pKa = 4.45SPGRR61 pKa = 11.84AVARR65 pKa = 11.84VLPVGTVIPTFVPGPDD81 pKa = 3.25RR82 pKa = 11.84NSDD85 pKa = 3.26KK86 pKa = 11.13LGYY89 pKa = 8.45CTGSTTMGPYY99 pKa = 10.16NLSTIGKK106 pKa = 8.71FALEE110 pKa = 4.01LAQLEE115 pKa = 4.58VHH117 pKa = 6.51FNSHH121 pKa = 7.46FILSFDD127 pKa = 3.97IPPDD131 pKa = 3.41FDD133 pKa = 4.89FYY135 pKa = 11.74NDD137 pKa = 3.67PPYY140 pKa = 10.91LQAHH144 pKa = 5.1YY145 pKa = 9.25TGKK148 pKa = 9.66PLKK151 pKa = 9.07TQVSLKK157 pKa = 10.5QVDD160 pKa = 3.62NMLFALSQLSFEE172 pKa = 4.41QTSFDD177 pKa = 4.75DD178 pKa = 4.5KK179 pKa = 10.78EE180 pKa = 3.9QTAIDD185 pKa = 3.92VSALGMATGSVAYY198 pKa = 9.98DD199 pKa = 3.01QTTNVSSIFKK209 pKa = 8.82WFQYY213 pKa = 8.63EE214 pKa = 4.41FKK216 pKa = 11.02APNDD220 pKa = 3.7KK221 pKa = 10.15IKK223 pKa = 10.71RR224 pKa = 11.84IISPDD229 pKa = 2.51IYY231 pKa = 9.82YY232 pKa = 10.82TMFFKK237 pKa = 10.51DD238 pKa = 3.77DD239 pKa = 3.76KK240 pKa = 11.58ADD242 pKa = 3.74GGVSYY247 pKa = 9.15ITLPNPGLQLFADD260 pKa = 3.51AVEE263 pKa = 4.27RR264 pKa = 11.84TEE266 pKa = 4.49VALNYY271 pKa = 8.79QLRR274 pKa = 11.84SGITVSHH281 pKa = 6.87IDD283 pKa = 3.18IFSNIPQASYY293 pKa = 11.29NSPLTPLFSFFHH305 pKa = 6.04SDD307 pKa = 3.71KK308 pKa = 11.01YY309 pKa = 11.07ILSSWLRR316 pKa = 11.84FQNYY320 pKa = 5.74YY321 pKa = 8.73TYY323 pKa = 10.86KK324 pKa = 10.27GRR326 pKa = 11.84NITAQEE332 pKa = 3.99LNYY335 pKa = 10.78GITEE339 pKa = 4.04DD340 pKa = 3.82MRR342 pKa = 11.84NLVYY346 pKa = 9.71VQCHH350 pKa = 5.68EE351 pKa = 4.12RR352 pKa = 11.84PEE354 pKa = 4.02MCKK357 pKa = 10.58CEE359 pKa = 4.18GNYY362 pKa = 10.24SSQALAIGAFGVDD375 pKa = 3.09STYY378 pKa = 11.25TKK380 pKa = 10.8VPNTAIYY387 pKa = 9.93EE388 pKa = 4.16NQTFQVHH395 pKa = 5.44SPILHH400 pKa = 6.2HH401 pKa = 6.6VGSVPLEE408 pKa = 4.07GPYY411 pKa = 10.3TLFSSALPDD420 pKa = 3.78LPDD423 pKa = 3.38IKK425 pKa = 10.88ASNLPAWLPEE435 pKa = 4.24SVGEE439 pKa = 4.14EE440 pKa = 3.78IEE442 pKa = 4.42GVLNAVDD449 pKa = 3.92NFYY452 pKa = 11.47KK453 pKa = 10.31HH454 pKa = 6.75VKK456 pKa = 10.16DD457 pKa = 4.12FWEE460 pKa = 4.1ILQHH464 pKa = 7.25IEE466 pKa = 4.64EE467 pKa = 4.37IVEE470 pKa = 4.1VLKK473 pKa = 11.03DD474 pKa = 3.45IASLNPFGGSTPDD487 pKa = 3.27AVPVV491 pKa = 3.51
Molecular weight: 55.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJW3|A0A1L3KJW3_9VIRU Uncharacterized protein OS=Beihai hepe-like virus 10 OX=1922381 PE=4 SV=1
MM1 pKa = 7.57SFTNFAGVPSEE12 pKa = 4.35EE13 pKa = 4.54KK14 pKa = 10.42LSEE17 pKa = 4.82LIFPTCPHH25 pKa = 6.58GEE27 pKa = 3.85ADD29 pKa = 4.08KK30 pKa = 11.51YY31 pKa = 10.7HH32 pKa = 6.78ALFNKK37 pKa = 8.55FQQINGGVSPIKK49 pKa = 10.06IDD51 pKa = 3.72AFCLGSAPKK60 pKa = 10.46QMWIAEE66 pKa = 4.16AYY68 pKa = 9.93YY69 pKa = 10.83NGLNFQGYY77 pKa = 9.33GSSKK81 pKa = 10.12SSAILEE87 pKa = 4.29VKK89 pKa = 9.64FQFMRR94 pKa = 11.84SVGQVDD100 pKa = 3.07SWANVGAAVSLDD112 pKa = 3.25SGTLKK117 pKa = 10.24TGTRR121 pKa = 11.84LVLTPFLNLRR131 pKa = 11.84VEE133 pKa = 4.52VFHH136 pKa = 6.61ATYY139 pKa = 10.06IYY141 pKa = 10.74LRR143 pKa = 11.84SNKK146 pKa = 9.86LYY148 pKa = 10.04YY149 pKa = 9.47LTAAGPSKK157 pKa = 10.46EE158 pKa = 3.92LAILFLRR165 pKa = 11.84SLLAA169 pKa = 4.53
MM1 pKa = 7.57SFTNFAGVPSEE12 pKa = 4.35EE13 pKa = 4.54KK14 pKa = 10.42LSEE17 pKa = 4.82LIFPTCPHH25 pKa = 6.58GEE27 pKa = 3.85ADD29 pKa = 4.08KK30 pKa = 11.51YY31 pKa = 10.7HH32 pKa = 6.78ALFNKK37 pKa = 8.55FQQINGGVSPIKK49 pKa = 10.06IDD51 pKa = 3.72AFCLGSAPKK60 pKa = 10.46QMWIAEE66 pKa = 4.16AYY68 pKa = 9.93YY69 pKa = 10.83NGLNFQGYY77 pKa = 9.33GSSKK81 pKa = 10.12SSAILEE87 pKa = 4.29VKK89 pKa = 9.64FQFMRR94 pKa = 11.84SVGQVDD100 pKa = 3.07SWANVGAAVSLDD112 pKa = 3.25SGTLKK117 pKa = 10.24TGTRR121 pKa = 11.84LVLTPFLNLRR131 pKa = 11.84VEE133 pKa = 4.52VFHH136 pKa = 6.61ATYY139 pKa = 10.06IYY141 pKa = 10.74LRR143 pKa = 11.84SNKK146 pKa = 9.86LYY148 pKa = 10.04YY149 pKa = 9.47LTAAGPSKK157 pKa = 10.46EE158 pKa = 3.92LAILFLRR165 pKa = 11.84SLLAA169 pKa = 4.53
Molecular weight: 18.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3549 |
146 |
2368 |
709.8 |
79.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.481 ± 0.587 | 1.662 ± 0.349 |
5.861 ± 0.617 | 7.411 ± 1.297 |
4.227 ± 0.825 | 5.889 ± 0.368 |
3.212 ± 0.59 | 5.917 ± 0.365 |
5.917 ± 0.876 | 7.608 ± 0.998 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.113 ± 0.275 | 4.367 ± 0.407 |
5.156 ± 0.426 | 4.142 ± 0.498 |
4.339 ± 0.716 | 6.114 ± 1.462 |
7.157 ± 0.773 | 5.466 ± 0.307 |
1.071 ± 0.172 | 3.888 ± 0.551 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
