
Leptolyngbyaceae cyanobacterium JSC-12
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Leptolyngbyaceae; unclassified Leptolyngbyaceae
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
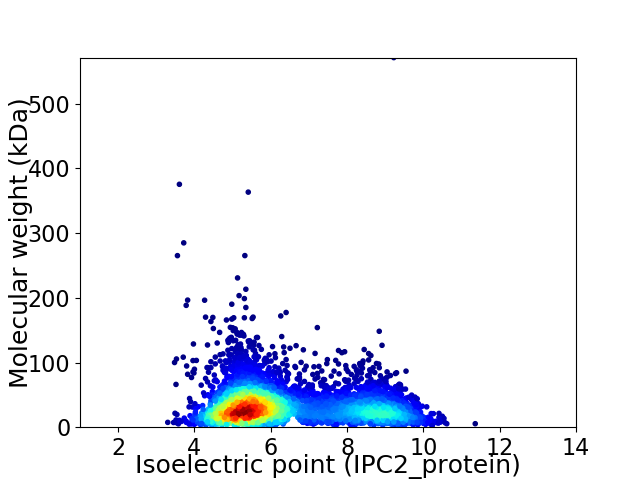
Virtual 2D-PAGE plot for 4673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
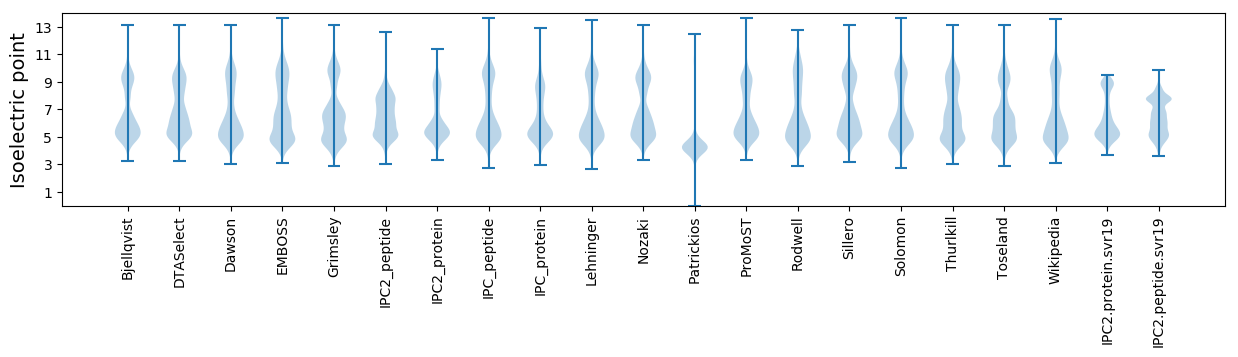
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K8GKS3|K8GKS3_9CYAN Uncharacterized protein OS=Leptolyngbyaceae cyanobacterium JSC-12 OX=864702 GN=OsccyDRAFT_2914 PE=4 SV=1
MM1 pKa = 7.55TSTSVEE7 pKa = 3.74QSAFAGLPSLLDD19 pKa = 3.46QSPLDD24 pKa = 3.6SSIPSLHH31 pKa = 6.08STNSAANLTPLQLPSFSGSSAILSVGNSIFTAVNIGALNGTVLGTGLVNSSDD83 pKa = 3.26PSDD86 pKa = 3.53YY87 pKa = 10.93FRR89 pKa = 11.84FTTSAPGSLSMSLTGMGTGDD109 pKa = 3.5ADD111 pKa = 3.28IYY113 pKa = 10.79LIQDD117 pKa = 3.36INNNGVLDD125 pKa = 4.0VNLGEE130 pKa = 4.24IIEE133 pKa = 4.35RR134 pKa = 11.84SRR136 pKa = 11.84SSSNTEE142 pKa = 3.68FLSVNLLAPGTYY154 pKa = 9.36YY155 pKa = 11.51ALVNHH160 pKa = 6.56FSGGTTNYY168 pKa = 10.34VLTLSADD175 pKa = 3.66TAGSNFLTARR185 pKa = 11.84NLGTLTASGSVSDD198 pKa = 4.43FVGNADD204 pKa = 3.64PSDD207 pKa = 3.94LYY209 pKa = 11.17RR210 pKa = 11.84FNLSNTSDD218 pKa = 3.44VTLTLSGLNNDD229 pKa = 3.52ADD231 pKa = 4.25LYY233 pKa = 11.09LIQDD237 pKa = 4.49LNQNGILDD245 pKa = 3.89NADD248 pKa = 3.42VLEE251 pKa = 3.88QSINFGSAAEE261 pKa = 4.51FIFAQGLAAGSYY273 pKa = 8.25FVQVAQYY280 pKa = 11.0SGDD283 pKa = 3.42TNYY286 pKa = 10.19TLTYY290 pKa = 10.7SVIPSDD296 pKa = 4.74AGNTLATATNLGTLAGRR313 pKa = 11.84RR314 pKa = 11.84AVNGSVSSTDD324 pKa = 3.16LVDD327 pKa = 3.83IYY329 pKa = 11.0RR330 pKa = 11.84FSTDD334 pKa = 3.21AISDD338 pKa = 3.49VRR340 pKa = 11.84LTLTGLSSDD349 pKa = 3.31ADD351 pKa = 3.72LYY353 pKa = 11.24LIQDD357 pKa = 3.87RR358 pKa = 11.84NNNGLIDD365 pKa = 4.05AGEE368 pKa = 4.23TIEE371 pKa = 4.5YY372 pKa = 10.03SILSGASSEE381 pKa = 4.75SINLSKK387 pKa = 10.8LAIGTYY393 pKa = 9.44FVAVEE398 pKa = 4.63HH399 pKa = 6.22YY400 pKa = 10.47SGSTNYY406 pKa = 9.86TLTLEE411 pKa = 4.53ADD413 pKa = 3.69SAGKK417 pKa = 10.18SLATARR423 pKa = 11.84NIGTLTSTRR432 pKa = 11.84TFNDD436 pKa = 3.7SISEE440 pKa = 3.88QDD442 pKa = 3.26SVDD445 pKa = 4.26FYY447 pKa = 11.05RR448 pKa = 11.84FNIIAPGNLTVRR460 pKa = 11.84LDD462 pKa = 3.26GMTADD467 pKa = 3.83GDD469 pKa = 4.03LYY471 pKa = 11.1LIRR474 pKa = 11.84DD475 pKa = 3.71INNNGLVEE483 pKa = 4.25INEE486 pKa = 4.4IIAVSATDD494 pKa = 5.04GISSEE499 pKa = 4.45TINFYY504 pKa = 11.29DD505 pKa = 4.23LAAGTYY511 pKa = 7.11YY512 pKa = 10.85VRR514 pKa = 11.84VNQYY518 pKa = 10.7SGNTNYY524 pKa = 10.68KK525 pKa = 10.25LSLTMEE531 pKa = 4.39PTRR534 pKa = 11.84PEE536 pKa = 4.29PGSTLTTATNLGTLSGRR553 pKa = 11.84RR554 pKa = 11.84SFSDD558 pKa = 3.4SLSLNDD564 pKa = 3.32VDD566 pKa = 5.0DD567 pKa = 5.29FYY569 pKa = 11.64RR570 pKa = 11.84FSLSTTSDD578 pKa = 3.34LQVALTGLTSNADD591 pKa = 3.37LFLIRR596 pKa = 11.84DD597 pKa = 3.68MNGNGIVDD605 pKa = 3.82SMDD608 pKa = 3.21VLQSSTSLGAVSEE621 pKa = 4.89FISLSGLAAGTYY633 pKa = 8.96FVGIRR638 pKa = 11.84GYY640 pKa = 11.8GNFTNYY646 pKa = 9.13TLSLTADD653 pKa = 3.5SAGEE657 pKa = 3.99TLATARR663 pKa = 11.84NIGVLTGSRR672 pKa = 11.84SFTDD676 pKa = 3.95FLSFQDD682 pKa = 4.34DD683 pKa = 3.28SDD685 pKa = 3.56IYY687 pKa = 10.84RR688 pKa = 11.84FSLNIMSNVSINLPGLTADD707 pKa = 3.71FDD709 pKa = 4.79LYY711 pKa = 11.06LGRR714 pKa = 11.84DD715 pKa = 3.55LNGNGLIDD723 pKa = 4.28PGEE726 pKa = 4.14TLAISATDD734 pKa = 3.82GSNSPEE740 pKa = 4.3SINISNLASGVYY752 pKa = 9.2YY753 pKa = 10.5VLVRR757 pKa = 11.84QFSGEE762 pKa = 3.83ANYY765 pKa = 9.43TLNLTATPVSS775 pKa = 3.7
MM1 pKa = 7.55TSTSVEE7 pKa = 3.74QSAFAGLPSLLDD19 pKa = 3.46QSPLDD24 pKa = 3.6SSIPSLHH31 pKa = 6.08STNSAANLTPLQLPSFSGSSAILSVGNSIFTAVNIGALNGTVLGTGLVNSSDD83 pKa = 3.26PSDD86 pKa = 3.53YY87 pKa = 10.93FRR89 pKa = 11.84FTTSAPGSLSMSLTGMGTGDD109 pKa = 3.5ADD111 pKa = 3.28IYY113 pKa = 10.79LIQDD117 pKa = 3.36INNNGVLDD125 pKa = 4.0VNLGEE130 pKa = 4.24IIEE133 pKa = 4.35RR134 pKa = 11.84SRR136 pKa = 11.84SSSNTEE142 pKa = 3.68FLSVNLLAPGTYY154 pKa = 9.36YY155 pKa = 11.51ALVNHH160 pKa = 6.56FSGGTTNYY168 pKa = 10.34VLTLSADD175 pKa = 3.66TAGSNFLTARR185 pKa = 11.84NLGTLTASGSVSDD198 pKa = 4.43FVGNADD204 pKa = 3.64PSDD207 pKa = 3.94LYY209 pKa = 11.17RR210 pKa = 11.84FNLSNTSDD218 pKa = 3.44VTLTLSGLNNDD229 pKa = 3.52ADD231 pKa = 4.25LYY233 pKa = 11.09LIQDD237 pKa = 4.49LNQNGILDD245 pKa = 3.89NADD248 pKa = 3.42VLEE251 pKa = 3.88QSINFGSAAEE261 pKa = 4.51FIFAQGLAAGSYY273 pKa = 8.25FVQVAQYY280 pKa = 11.0SGDD283 pKa = 3.42TNYY286 pKa = 10.19TLTYY290 pKa = 10.7SVIPSDD296 pKa = 4.74AGNTLATATNLGTLAGRR313 pKa = 11.84RR314 pKa = 11.84AVNGSVSSTDD324 pKa = 3.16LVDD327 pKa = 3.83IYY329 pKa = 11.0RR330 pKa = 11.84FSTDD334 pKa = 3.21AISDD338 pKa = 3.49VRR340 pKa = 11.84LTLTGLSSDD349 pKa = 3.31ADD351 pKa = 3.72LYY353 pKa = 11.24LIQDD357 pKa = 3.87RR358 pKa = 11.84NNNGLIDD365 pKa = 4.05AGEE368 pKa = 4.23TIEE371 pKa = 4.5YY372 pKa = 10.03SILSGASSEE381 pKa = 4.75SINLSKK387 pKa = 10.8LAIGTYY393 pKa = 9.44FVAVEE398 pKa = 4.63HH399 pKa = 6.22YY400 pKa = 10.47SGSTNYY406 pKa = 9.86TLTLEE411 pKa = 4.53ADD413 pKa = 3.69SAGKK417 pKa = 10.18SLATARR423 pKa = 11.84NIGTLTSTRR432 pKa = 11.84TFNDD436 pKa = 3.7SISEE440 pKa = 3.88QDD442 pKa = 3.26SVDD445 pKa = 4.26FYY447 pKa = 11.05RR448 pKa = 11.84FNIIAPGNLTVRR460 pKa = 11.84LDD462 pKa = 3.26GMTADD467 pKa = 3.83GDD469 pKa = 4.03LYY471 pKa = 11.1LIRR474 pKa = 11.84DD475 pKa = 3.71INNNGLVEE483 pKa = 4.25INEE486 pKa = 4.4IIAVSATDD494 pKa = 5.04GISSEE499 pKa = 4.45TINFYY504 pKa = 11.29DD505 pKa = 4.23LAAGTYY511 pKa = 7.11YY512 pKa = 10.85VRR514 pKa = 11.84VNQYY518 pKa = 10.7SGNTNYY524 pKa = 10.68KK525 pKa = 10.25LSLTMEE531 pKa = 4.39PTRR534 pKa = 11.84PEE536 pKa = 4.29PGSTLTTATNLGTLSGRR553 pKa = 11.84RR554 pKa = 11.84SFSDD558 pKa = 3.4SLSLNDD564 pKa = 3.32VDD566 pKa = 5.0DD567 pKa = 5.29FYY569 pKa = 11.64RR570 pKa = 11.84FSLSTTSDD578 pKa = 3.34LQVALTGLTSNADD591 pKa = 3.37LFLIRR596 pKa = 11.84DD597 pKa = 3.68MNGNGIVDD605 pKa = 3.82SMDD608 pKa = 3.21VLQSSTSLGAVSEE621 pKa = 4.89FISLSGLAAGTYY633 pKa = 8.96FVGIRR638 pKa = 11.84GYY640 pKa = 11.8GNFTNYY646 pKa = 9.13TLSLTADD653 pKa = 3.5SAGEE657 pKa = 3.99TLATARR663 pKa = 11.84NIGVLTGSRR672 pKa = 11.84SFTDD676 pKa = 3.95FLSFQDD682 pKa = 4.34DD683 pKa = 3.28SDD685 pKa = 3.56IYY687 pKa = 10.84RR688 pKa = 11.84FSLNIMSNVSINLPGLTADD707 pKa = 3.71FDD709 pKa = 4.79LYY711 pKa = 11.06LGRR714 pKa = 11.84DD715 pKa = 3.55LNGNGLIDD723 pKa = 4.28PGEE726 pKa = 4.14TLAISATDD734 pKa = 3.82GSNSPEE740 pKa = 4.3SINISNLASGVYY752 pKa = 9.2YY753 pKa = 10.5VLVRR757 pKa = 11.84QFSGEE762 pKa = 3.83ANYY765 pKa = 9.43TLNLTATPVSS775 pKa = 3.7
Molecular weight: 81.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K8GLJ1|K8GLJ1_9CYAN Light-independent protochlorophyllide reductase subunit B OS=Leptolyngbyaceae cyanobacterium JSC-12 OX=864702 GN=chlB PE=3 SV=1
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 9.1RR13 pKa = 11.84KK14 pKa = 7.65RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.18NGRR29 pKa = 11.84RR30 pKa = 11.84VLKK33 pKa = 10.17ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.47GRR40 pKa = 11.84ARR42 pKa = 11.84LAVTT46 pKa = 4.38
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 9.1RR13 pKa = 11.84KK14 pKa = 7.65RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.18NGRR29 pKa = 11.84RR30 pKa = 11.84VLKK33 pKa = 10.17ARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.47GRR40 pKa = 11.84ARR42 pKa = 11.84LAVTT46 pKa = 4.38
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1516644 |
23 |
5284 |
324.6 |
35.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.923 ± 0.038 | 0.989 ± 0.013 |
4.923 ± 0.034 | 5.834 ± 0.04 |
3.885 ± 0.025 | 6.832 ± 0.046 |
1.966 ± 0.022 | 6.051 ± 0.03 |
3.818 ± 0.033 | 11.153 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.961 ± 0.016 | 3.757 ± 0.031 |
5.178 ± 0.029 | 5.45 ± 0.036 |
5.716 ± 0.03 | 6.397 ± 0.036 |
5.887 ± 0.034 | 6.974 ± 0.028 |
1.456 ± 0.018 | 2.851 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
