
Bombyx mori densovirus Zhenjiang
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Mouviricetes; Polivirales; Bidnaviridae; Bidensovirus; Bombyx mori bidensovirus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
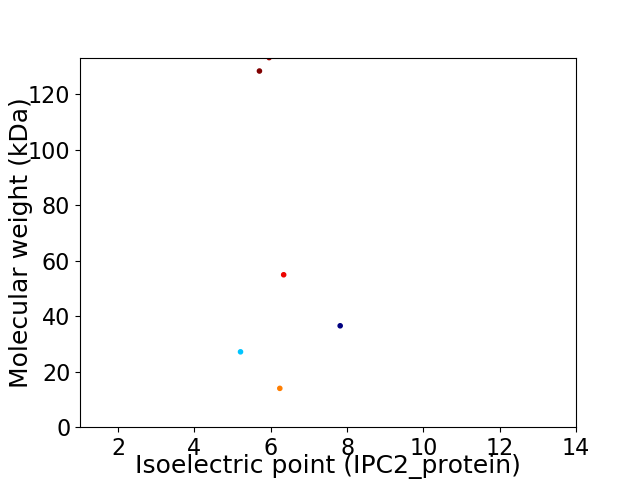
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2ZA91|B2ZA91_9VIRU Nonstructural protein OS=Bombyx mori densovirus Zhenjiang OX=530042 PE=4 SV=1
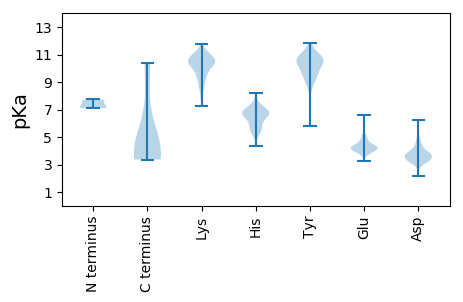
MM1 pKa = 7.68LRR3 pKa = 11.84LTDD6 pKa = 3.46LHH8 pKa = 7.07KK9 pKa = 10.59PSCEE13 pKa = 3.81PKK15 pKa = 10.08GLSFYY20 pKa = 10.52AYY22 pKa = 9.68KK23 pKa = 10.92AFIPYY28 pKa = 9.49RR29 pKa = 11.84EE30 pKa = 4.2KK31 pKa = 11.0LSKK34 pKa = 9.85EE35 pKa = 3.93WNKK38 pKa = 10.6LPVCIKK44 pKa = 10.47DD45 pKa = 3.33QEE47 pKa = 4.08VDD49 pKa = 2.71IKK51 pKa = 11.27QEE53 pKa = 3.96VKK55 pKa = 10.36EE56 pKa = 4.15DD57 pKa = 3.26QLIRR61 pKa = 11.84KK62 pKa = 8.42IYY64 pKa = 9.82KK65 pKa = 7.97WDD67 pKa = 3.57SVFDD71 pKa = 3.34SWNWSKK77 pKa = 11.09GYY79 pKa = 10.4KK80 pKa = 9.44LPKK83 pKa = 9.34PLYY86 pKa = 8.65EE87 pKa = 4.17LLEE90 pKa = 5.14IYY92 pKa = 9.31MKK94 pKa = 10.49LWKK97 pKa = 10.02LDD99 pKa = 3.26NRR101 pKa = 11.84TTYY104 pKa = 10.33PDD106 pKa = 3.06WGEE109 pKa = 3.52SSMIFKK115 pKa = 10.3CYY117 pKa = 10.02RR118 pKa = 11.84YY119 pKa = 9.71IRR121 pKa = 11.84NKK123 pKa = 10.29RR124 pKa = 11.84MEE126 pKa = 5.19DD127 pKa = 3.11IEE129 pKa = 4.24NLSTEE134 pKa = 4.36EE135 pKa = 4.17FNDD138 pKa = 3.75SNFNGWPKK146 pKa = 9.85TMWYY150 pKa = 9.25ICRR153 pKa = 11.84NCFDD157 pKa = 3.74NGIVINNNSNDD168 pKa = 3.38YY169 pKa = 11.15YY170 pKa = 8.68YY171 pKa = 10.78TCDD174 pKa = 3.9FSDD177 pKa = 3.81NPTLLHH183 pKa = 5.17YY184 pKa = 9.59TDD186 pKa = 3.71YY187 pKa = 10.99FYY189 pKa = 11.47YY190 pKa = 10.21ILNNDD195 pKa = 3.66YY196 pKa = 9.93WCYY199 pKa = 9.85DD200 pKa = 3.53CKK202 pKa = 10.4YY203 pKa = 8.8TPLFTLTTLFCLEE216 pKa = 4.88SINDD220 pKa = 3.74SEE222 pKa = 4.42
MM1 pKa = 7.68LRR3 pKa = 11.84LTDD6 pKa = 3.46LHH8 pKa = 7.07KK9 pKa = 10.59PSCEE13 pKa = 3.81PKK15 pKa = 10.08GLSFYY20 pKa = 10.52AYY22 pKa = 9.68KK23 pKa = 10.92AFIPYY28 pKa = 9.49RR29 pKa = 11.84EE30 pKa = 4.2KK31 pKa = 11.0LSKK34 pKa = 9.85EE35 pKa = 3.93WNKK38 pKa = 10.6LPVCIKK44 pKa = 10.47DD45 pKa = 3.33QEE47 pKa = 4.08VDD49 pKa = 2.71IKK51 pKa = 11.27QEE53 pKa = 3.96VKK55 pKa = 10.36EE56 pKa = 4.15DD57 pKa = 3.26QLIRR61 pKa = 11.84KK62 pKa = 8.42IYY64 pKa = 9.82KK65 pKa = 7.97WDD67 pKa = 3.57SVFDD71 pKa = 3.34SWNWSKK77 pKa = 11.09GYY79 pKa = 10.4KK80 pKa = 9.44LPKK83 pKa = 9.34PLYY86 pKa = 8.65EE87 pKa = 4.17LLEE90 pKa = 5.14IYY92 pKa = 9.31MKK94 pKa = 10.49LWKK97 pKa = 10.02LDD99 pKa = 3.26NRR101 pKa = 11.84TTYY104 pKa = 10.33PDD106 pKa = 3.06WGEE109 pKa = 3.52SSMIFKK115 pKa = 10.3CYY117 pKa = 10.02RR118 pKa = 11.84YY119 pKa = 9.71IRR121 pKa = 11.84NKK123 pKa = 10.29RR124 pKa = 11.84MEE126 pKa = 5.19DD127 pKa = 3.11IEE129 pKa = 4.24NLSTEE134 pKa = 4.36EE135 pKa = 4.17FNDD138 pKa = 3.75SNFNGWPKK146 pKa = 9.85TMWYY150 pKa = 9.25ICRR153 pKa = 11.84NCFDD157 pKa = 3.74NGIVINNNSNDD168 pKa = 3.38YY169 pKa = 11.15YY170 pKa = 8.68YY171 pKa = 10.78TCDD174 pKa = 3.9FSDD177 pKa = 3.81NPTLLHH183 pKa = 5.17YY184 pKa = 9.59TDD186 pKa = 3.71YY187 pKa = 10.99FYY189 pKa = 11.47YY190 pKa = 10.21ILNNDD195 pKa = 3.66YY196 pKa = 9.93WCYY199 pKa = 9.85DD200 pKa = 3.53CKK202 pKa = 10.4YY203 pKa = 8.8TPLFTLTTLFCLEE216 pKa = 4.88SINDD220 pKa = 3.74SEE222 pKa = 4.42
Molecular weight: 27.14 kDa
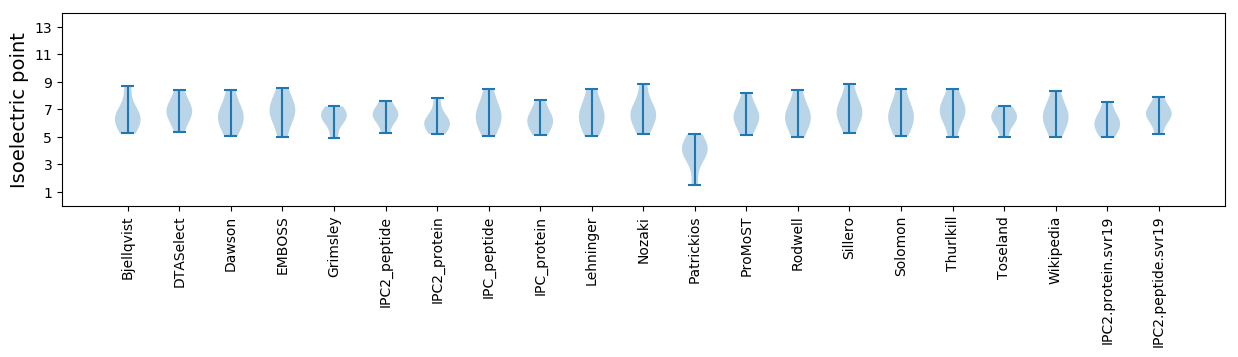
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2ZA88|B2ZA88_9VIRU Nonstructural protein OS=Bombyx mori densovirus Zhenjiang OX=530042 PE=4 SV=1
MM1 pKa = 7.46EE2 pKa = 5.22SKK4 pKa = 11.13SNFRR8 pKa = 11.84ILSNEE13 pKa = 4.18IISKK17 pKa = 9.68PSSLISNISSPSTSLIPRR35 pKa = 11.84SLQKK39 pKa = 9.98SAKK42 pKa = 9.2RR43 pKa = 11.84LTISSLEE50 pKa = 4.18SGNGKK55 pKa = 7.88QQMKK59 pKa = 10.59SKK61 pKa = 10.17LAQVQRR67 pKa = 11.84ALRR70 pKa = 11.84FCRR73 pKa = 11.84DD74 pKa = 3.45YY75 pKa = 11.6NIVKK79 pKa = 9.91YY80 pKa = 9.12YY81 pKa = 10.43QFMDD85 pKa = 3.96IPPEE89 pKa = 3.9EE90 pKa = 3.99YY91 pKa = 10.06RR92 pKa = 11.84EE93 pKa = 4.09LRR95 pKa = 11.84THH97 pKa = 6.91CSKK100 pKa = 11.15DD101 pKa = 3.61VLEE104 pKa = 4.48TAADD108 pKa = 3.38INYY111 pKa = 9.33QEE113 pKa = 4.37YY114 pKa = 10.55YY115 pKa = 10.04YY116 pKa = 10.98EE117 pKa = 4.15EE118 pKa = 4.36VNNRR122 pKa = 11.84LQVFLSYY129 pKa = 11.65GDD131 pKa = 4.1GSWGEE136 pKa = 4.08EE137 pKa = 3.85EE138 pKa = 4.68EE139 pKa = 4.48EE140 pKa = 3.62IVMLFKK146 pKa = 11.02HH147 pKa = 5.96NNVDD151 pKa = 2.84ISKK154 pKa = 9.91FCEE157 pKa = 3.51ALYY160 pKa = 10.81SIFTMSDD167 pKa = 2.96YY168 pKa = 11.07RR169 pKa = 11.84KK170 pKa = 10.24NCIFLNGNIKK180 pKa = 9.09TGKK183 pKa = 7.22TLMMRR188 pKa = 11.84LITGNFKK195 pKa = 10.87CVGKK199 pKa = 10.72GCNNGVNSEE208 pKa = 4.29FYY210 pKa = 8.22YY211 pKa = 10.72QKK213 pKa = 10.81FIGRR217 pKa = 11.84SICVLEE223 pKa = 4.66EE224 pKa = 4.26PFLDD228 pKa = 4.18PQTLEE233 pKa = 3.98DD234 pKa = 4.2FKK236 pKa = 11.43SIFGGEE242 pKa = 4.25GFDD245 pKa = 4.68ISCKK249 pKa = 9.13FHH251 pKa = 7.23NPQTLNRR258 pKa = 11.84TPFIITSNFSLLSRR272 pKa = 11.84GHH274 pKa = 6.59APSISEE280 pKa = 3.89MAIKK284 pKa = 10.53DD285 pKa = 3.24RR286 pKa = 11.84FYY288 pKa = 10.79IFNFNNSYY296 pKa = 9.09TPKK299 pKa = 10.55CKK301 pKa = 10.33LSPKK305 pKa = 10.21NFVNVYY311 pKa = 9.23NKK313 pKa = 10.75YY314 pKa = 10.39YY315 pKa = 10.78GG316 pKa = 3.37
MM1 pKa = 7.46EE2 pKa = 5.22SKK4 pKa = 11.13SNFRR8 pKa = 11.84ILSNEE13 pKa = 4.18IISKK17 pKa = 9.68PSSLISNISSPSTSLIPRR35 pKa = 11.84SLQKK39 pKa = 9.98SAKK42 pKa = 9.2RR43 pKa = 11.84LTISSLEE50 pKa = 4.18SGNGKK55 pKa = 7.88QQMKK59 pKa = 10.59SKK61 pKa = 10.17LAQVQRR67 pKa = 11.84ALRR70 pKa = 11.84FCRR73 pKa = 11.84DD74 pKa = 3.45YY75 pKa = 11.6NIVKK79 pKa = 9.91YY80 pKa = 9.12YY81 pKa = 10.43QFMDD85 pKa = 3.96IPPEE89 pKa = 3.9EE90 pKa = 3.99YY91 pKa = 10.06RR92 pKa = 11.84EE93 pKa = 4.09LRR95 pKa = 11.84THH97 pKa = 6.91CSKK100 pKa = 11.15DD101 pKa = 3.61VLEE104 pKa = 4.48TAADD108 pKa = 3.38INYY111 pKa = 9.33QEE113 pKa = 4.37YY114 pKa = 10.55YY115 pKa = 10.04YY116 pKa = 10.98EE117 pKa = 4.15EE118 pKa = 4.36VNNRR122 pKa = 11.84LQVFLSYY129 pKa = 11.65GDD131 pKa = 4.1GSWGEE136 pKa = 4.08EE137 pKa = 3.85EE138 pKa = 4.68EE139 pKa = 4.48EE140 pKa = 3.62IVMLFKK146 pKa = 11.02HH147 pKa = 5.96NNVDD151 pKa = 2.84ISKK154 pKa = 9.91FCEE157 pKa = 3.51ALYY160 pKa = 10.81SIFTMSDD167 pKa = 2.96YY168 pKa = 11.07RR169 pKa = 11.84KK170 pKa = 10.24NCIFLNGNIKK180 pKa = 9.09TGKK183 pKa = 7.22TLMMRR188 pKa = 11.84LITGNFKK195 pKa = 10.87CVGKK199 pKa = 10.72GCNNGVNSEE208 pKa = 4.29FYY210 pKa = 8.22YY211 pKa = 10.72QKK213 pKa = 10.81FIGRR217 pKa = 11.84SICVLEE223 pKa = 4.66EE224 pKa = 4.26PFLDD228 pKa = 4.18PQTLEE233 pKa = 3.98DD234 pKa = 4.2FKK236 pKa = 11.43SIFGGEE242 pKa = 4.25GFDD245 pKa = 4.68ISCKK249 pKa = 9.13FHH251 pKa = 7.23NPQTLNRR258 pKa = 11.84TPFIITSNFSLLSRR272 pKa = 11.84GHH274 pKa = 6.59APSISEE280 pKa = 3.89MAIKK284 pKa = 10.53DD285 pKa = 3.24RR286 pKa = 11.84FYY288 pKa = 10.79IFNFNNSYY296 pKa = 9.09TPKK299 pKa = 10.55CKK301 pKa = 10.33LSPKK305 pKa = 10.21NFVNVYY311 pKa = 9.23NKK313 pKa = 10.75YY314 pKa = 10.39YY315 pKa = 10.78GG316 pKa = 3.37
Molecular weight: 36.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3438 |
126 |
1160 |
573.0 |
65.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.363 ± 0.566 | 1.687 ± 0.334 |
6.137 ± 0.65 | 5.526 ± 0.455 |
5.061 ± 0.45 | 5.177 ± 0.985 |
1.483 ± 0.373 | 8.581 ± 0.629 |
6.457 ± 1.241 | 8.377 ± 0.617 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.571 ± 0.23 | 8.173 ± 0.643 |
4.101 ± 0.247 | 3.229 ± 0.723 |
4.654 ± 0.776 | 7.533 ± 0.916 |
6.457 ± 0.499 | 4.945 ± 0.341 |
1.105 ± 0.306 | 5.381 ± 0.591 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
