
Beihai hermit crab virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Chuviridae; Mivirus; Hermit mivirus
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
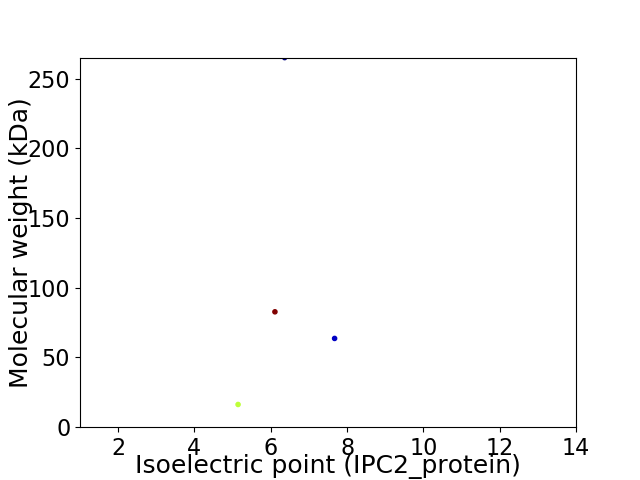
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMV0|A0A1L3KMV0_9VIRU Large structural protein OS=Beihai hermit crab virus 3 OX=1922390 PE=4 SV=1
MM1 pKa = 7.46SLALLEE7 pKa = 4.62RR8 pKa = 11.84IARR11 pKa = 11.84LEE13 pKa = 4.15AEE15 pKa = 4.39VSRR18 pKa = 11.84LKK20 pKa = 11.0LQLDD24 pKa = 3.65KK25 pKa = 10.64MRR27 pKa = 11.84KK28 pKa = 7.43SEE30 pKa = 4.33RR31 pKa = 11.84EE32 pKa = 4.04STHH35 pKa = 5.36HH36 pKa = 6.18LRR38 pKa = 11.84RR39 pKa = 11.84STRR42 pKa = 11.84QVDD45 pKa = 3.56RR46 pKa = 11.84DD47 pKa = 3.81LLTLAKK53 pKa = 10.33QEE55 pKa = 4.24TLSEE59 pKa = 4.43GGSGGRR65 pKa = 11.84GQILNVYY72 pKa = 9.17EE73 pKa = 4.92DD74 pKa = 5.16LPTSHH79 pKa = 7.83PKK81 pKa = 9.25PQSSFGPGEE90 pKa = 4.19GFSPPKK96 pKa = 10.82GEE98 pKa = 4.03IQPEE102 pKa = 4.11PSCPDD107 pKa = 3.5EE108 pKa = 4.47EE109 pKa = 4.49PKK111 pKa = 10.7EE112 pKa = 4.3SPEE115 pKa = 4.06LVGVTAPPTVDD126 pKa = 2.83IAPLPEE132 pKa = 4.69PFDD135 pKa = 3.77PVGDD139 pKa = 3.9SFAALLAKK147 pKa = 10.38FNN149 pKa = 4.18
MM1 pKa = 7.46SLALLEE7 pKa = 4.62RR8 pKa = 11.84IARR11 pKa = 11.84LEE13 pKa = 4.15AEE15 pKa = 4.39VSRR18 pKa = 11.84LKK20 pKa = 11.0LQLDD24 pKa = 3.65KK25 pKa = 10.64MRR27 pKa = 11.84KK28 pKa = 7.43SEE30 pKa = 4.33RR31 pKa = 11.84EE32 pKa = 4.04STHH35 pKa = 5.36HH36 pKa = 6.18LRR38 pKa = 11.84RR39 pKa = 11.84STRR42 pKa = 11.84QVDD45 pKa = 3.56RR46 pKa = 11.84DD47 pKa = 3.81LLTLAKK53 pKa = 10.33QEE55 pKa = 4.24TLSEE59 pKa = 4.43GGSGGRR65 pKa = 11.84GQILNVYY72 pKa = 9.17EE73 pKa = 4.92DD74 pKa = 5.16LPTSHH79 pKa = 7.83PKK81 pKa = 9.25PQSSFGPGEE90 pKa = 4.19GFSPPKK96 pKa = 10.82GEE98 pKa = 4.03IQPEE102 pKa = 4.11PSCPDD107 pKa = 3.5EE108 pKa = 4.47EE109 pKa = 4.49PKK111 pKa = 10.7EE112 pKa = 4.3SPEE115 pKa = 4.06LVGVTAPPTVDD126 pKa = 2.83IAPLPEE132 pKa = 4.69PFDD135 pKa = 3.77PVGDD139 pKa = 3.9SFAALLAKK147 pKa = 10.38FNN149 pKa = 4.18
Molecular weight: 16.29 kDa
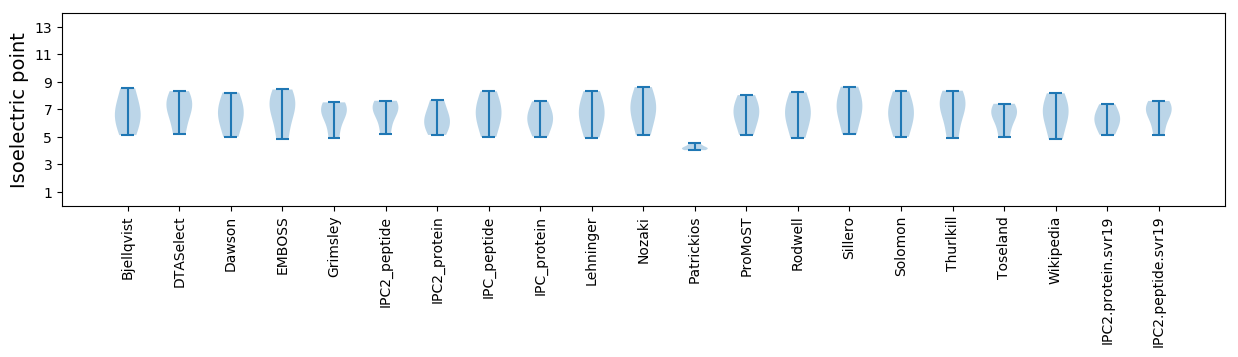
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMP0|A0A1L3KMP0_9VIRU Putative glycoprotein OS=Beihai hermit crab virus 3 OX=1922390 PE=4 SV=1
MM1 pKa = 7.38SRR3 pKa = 11.84YY4 pKa = 9.14FRR6 pKa = 11.84ANHH9 pKa = 5.29QAQKK13 pKa = 10.62ASNVAVAAGATGVVGTAFGWAQHH36 pKa = 5.43QADD39 pKa = 3.95AVSLKK44 pKa = 10.81GSLSHH49 pKa = 7.7LAQDD53 pKa = 3.81YY54 pKa = 8.48MDD56 pKa = 3.88SVRR59 pKa = 11.84PEE61 pKa = 3.41TDD63 pKa = 3.39FRR65 pKa = 11.84ILQALAIHH73 pKa = 6.07MAEE76 pKa = 4.06LWVKK80 pKa = 10.35SKK82 pKa = 10.64EE83 pKa = 4.16FEE85 pKa = 4.39SPHH88 pKa = 7.84LDD90 pKa = 3.45AEE92 pKa = 4.25NLAYY96 pKa = 10.6VHH98 pKa = 6.89LAATLFCRR106 pKa = 11.84WSAPGAVSLADD117 pKa = 3.37YY118 pKa = 10.8HH119 pKa = 7.03RR120 pKa = 11.84LLSMAGGALTPAQKK134 pKa = 9.77ATIQARR140 pKa = 11.84IATAMNASHH149 pKa = 7.66DD150 pKa = 4.17GNGAWVAATAVTKK163 pKa = 10.64GKK165 pKa = 10.21YY166 pKa = 9.37RR167 pKa = 11.84DD168 pKa = 3.38VGGQGLHH175 pKa = 6.22FAANTMPHH183 pKa = 6.98RR184 pKa = 11.84IHH186 pKa = 6.95RR187 pKa = 11.84HH188 pKa = 4.26LLEE191 pKa = 4.48IAAGTAANPPAVVQANDD208 pKa = 3.33IDD210 pKa = 3.65QHH212 pKa = 7.65LIADD216 pKa = 3.64NTYY219 pKa = 10.91SVNAQLFYY227 pKa = 10.48HH228 pKa = 5.41QCVHH232 pKa = 6.89NIATAGAKK240 pKa = 7.59QRR242 pKa = 11.84KK243 pKa = 8.08MYY245 pKa = 10.72ACALMLFVSSFVRR258 pKa = 11.84TGAPTNQWVRR268 pKa = 11.84KK269 pKa = 8.26RR270 pKa = 11.84AAGLEE275 pKa = 4.07EE276 pKa = 4.37GMGLATGEE284 pKa = 4.21IEE286 pKa = 3.78IHH288 pKa = 5.68SAAFEE293 pKa = 4.29TFRR296 pKa = 11.84SLVGIKK302 pKa = 10.3DD303 pKa = 3.31VTQAQTHH310 pKa = 5.58SALSLFATHH319 pKa = 7.42ADD321 pKa = 3.5TLGVPRR327 pKa = 11.84IRR329 pKa = 11.84WMVEE333 pKa = 3.11QAAYY337 pKa = 9.86TNMTIPVTLAQVLLQYY353 pKa = 11.06DD354 pKa = 4.4YY355 pKa = 11.19IPYY358 pKa = 10.52GLLFRR363 pKa = 11.84DD364 pKa = 3.84VTQEE368 pKa = 3.46LAALVKK374 pKa = 9.95ATVAAYY380 pKa = 9.05LNPYY384 pKa = 9.49IGLHH388 pKa = 4.66SVKK391 pKa = 10.3FPSTKK396 pKa = 9.98FATIATATVEE406 pKa = 3.97ICRR409 pKa = 11.84EE410 pKa = 3.59IGMTTFDD417 pKa = 3.95RR418 pKa = 11.84YY419 pKa = 10.42KK420 pKa = 10.7GVYY423 pKa = 9.34SGCDD427 pKa = 3.1QNKK430 pKa = 9.53KK431 pKa = 10.69DD432 pKa = 4.15NIVAAFRR439 pKa = 11.84SIRR442 pKa = 11.84QDD444 pKa = 2.82SSASNLVAEE453 pKa = 4.69AMSAFMSGTVPTITVRR469 pKa = 11.84RR470 pKa = 11.84GVMNVDD476 pKa = 3.54YY477 pKa = 9.34STAAAAVALPANWDD491 pKa = 3.65ANFKK495 pKa = 10.12NLCTNHH501 pKa = 6.67RR502 pKa = 11.84VPAGGIGTFEE512 pKa = 3.33IRR514 pKa = 11.84ARR516 pKa = 11.84DD517 pKa = 3.72LYY519 pKa = 11.5AFLKK523 pKa = 8.72TAEE526 pKa = 4.21RR527 pKa = 11.84ANPNTNAAILNYY539 pKa = 8.55YY540 pKa = 8.51TVLEE544 pKa = 4.36TCGVDD549 pKa = 3.99LQTPPLDD556 pKa = 3.51NAFSVASFRR565 pKa = 11.84RR566 pKa = 11.84ADD568 pKa = 3.72PAVGTPLHH576 pKa = 6.98AYY578 pKa = 7.59MAANGDD584 pKa = 3.63RR585 pKa = 11.84AGFEE589 pKa = 4.09
MM1 pKa = 7.38SRR3 pKa = 11.84YY4 pKa = 9.14FRR6 pKa = 11.84ANHH9 pKa = 5.29QAQKK13 pKa = 10.62ASNVAVAAGATGVVGTAFGWAQHH36 pKa = 5.43QADD39 pKa = 3.95AVSLKK44 pKa = 10.81GSLSHH49 pKa = 7.7LAQDD53 pKa = 3.81YY54 pKa = 8.48MDD56 pKa = 3.88SVRR59 pKa = 11.84PEE61 pKa = 3.41TDD63 pKa = 3.39FRR65 pKa = 11.84ILQALAIHH73 pKa = 6.07MAEE76 pKa = 4.06LWVKK80 pKa = 10.35SKK82 pKa = 10.64EE83 pKa = 4.16FEE85 pKa = 4.39SPHH88 pKa = 7.84LDD90 pKa = 3.45AEE92 pKa = 4.25NLAYY96 pKa = 10.6VHH98 pKa = 6.89LAATLFCRR106 pKa = 11.84WSAPGAVSLADD117 pKa = 3.37YY118 pKa = 10.8HH119 pKa = 7.03RR120 pKa = 11.84LLSMAGGALTPAQKK134 pKa = 9.77ATIQARR140 pKa = 11.84IATAMNASHH149 pKa = 7.66DD150 pKa = 4.17GNGAWVAATAVTKK163 pKa = 10.64GKK165 pKa = 10.21YY166 pKa = 9.37RR167 pKa = 11.84DD168 pKa = 3.38VGGQGLHH175 pKa = 6.22FAANTMPHH183 pKa = 6.98RR184 pKa = 11.84IHH186 pKa = 6.95RR187 pKa = 11.84HH188 pKa = 4.26LLEE191 pKa = 4.48IAAGTAANPPAVVQANDD208 pKa = 3.33IDD210 pKa = 3.65QHH212 pKa = 7.65LIADD216 pKa = 3.64NTYY219 pKa = 10.91SVNAQLFYY227 pKa = 10.48HH228 pKa = 5.41QCVHH232 pKa = 6.89NIATAGAKK240 pKa = 7.59QRR242 pKa = 11.84KK243 pKa = 8.08MYY245 pKa = 10.72ACALMLFVSSFVRR258 pKa = 11.84TGAPTNQWVRR268 pKa = 11.84KK269 pKa = 8.26RR270 pKa = 11.84AAGLEE275 pKa = 4.07EE276 pKa = 4.37GMGLATGEE284 pKa = 4.21IEE286 pKa = 3.78IHH288 pKa = 5.68SAAFEE293 pKa = 4.29TFRR296 pKa = 11.84SLVGIKK302 pKa = 10.3DD303 pKa = 3.31VTQAQTHH310 pKa = 5.58SALSLFATHH319 pKa = 7.42ADD321 pKa = 3.5TLGVPRR327 pKa = 11.84IRR329 pKa = 11.84WMVEE333 pKa = 3.11QAAYY337 pKa = 9.86TNMTIPVTLAQVLLQYY353 pKa = 11.06DD354 pKa = 4.4YY355 pKa = 11.19IPYY358 pKa = 10.52GLLFRR363 pKa = 11.84DD364 pKa = 3.84VTQEE368 pKa = 3.46LAALVKK374 pKa = 9.95ATVAAYY380 pKa = 9.05LNPYY384 pKa = 9.49IGLHH388 pKa = 4.66SVKK391 pKa = 10.3FPSTKK396 pKa = 9.98FATIATATVEE406 pKa = 3.97ICRR409 pKa = 11.84EE410 pKa = 3.59IGMTTFDD417 pKa = 3.95RR418 pKa = 11.84YY419 pKa = 10.42KK420 pKa = 10.7GVYY423 pKa = 9.34SGCDD427 pKa = 3.1QNKK430 pKa = 9.53KK431 pKa = 10.69DD432 pKa = 4.15NIVAAFRR439 pKa = 11.84SIRR442 pKa = 11.84QDD444 pKa = 2.82SSASNLVAEE453 pKa = 4.69AMSAFMSGTVPTITVRR469 pKa = 11.84RR470 pKa = 11.84GVMNVDD476 pKa = 3.54YY477 pKa = 9.34STAAAAVALPANWDD491 pKa = 3.65ANFKK495 pKa = 10.12NLCTNHH501 pKa = 6.67RR502 pKa = 11.84VPAGGIGTFEE512 pKa = 3.33IRR514 pKa = 11.84ARR516 pKa = 11.84DD517 pKa = 3.72LYY519 pKa = 11.5AFLKK523 pKa = 8.72TAEE526 pKa = 4.21RR527 pKa = 11.84ANPNTNAAILNYY539 pKa = 8.55YY540 pKa = 8.51TVLEE544 pKa = 4.36TCGVDD549 pKa = 3.99LQTPPLDD556 pKa = 3.51NAFSVASFRR565 pKa = 11.84RR566 pKa = 11.84ADD568 pKa = 3.72PAVGTPLHH576 pKa = 6.98AYY578 pKa = 7.59MAANGDD584 pKa = 3.63RR585 pKa = 11.84AGFEE589 pKa = 4.09
Molecular weight: 63.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3805 |
149 |
2328 |
951.3 |
106.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.464 ± 2.694 | 1.656 ± 0.211 |
4.941 ± 0.245 | 4.993 ± 0.718 |
4.047 ± 0.059 | 6.045 ± 0.394 |
3.627 ± 0.558 | 5.861 ± 0.758 |
3.653 ± 0.534 | 11.353 ± 1.245 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.628 ± 0.273 | 3.89 ± 0.349 |
5.151 ± 0.552 | 4.678 ± 0.258 |
5.782 ± 0.458 | 7.227 ± 1.548 |
6.15 ± 0.48 | 6.018 ± 0.365 |
1.945 ± 0.369 | 2.891 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
