
Olsenella uli (strain ATCC 49627 / DSM 7084 / CIP 109912 / JCM 12494 / NCIMB 702895 / VPI D76D-27C) (Lactobacillus uli)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Atopobiaceae; Olsenella; Olsenella uli
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
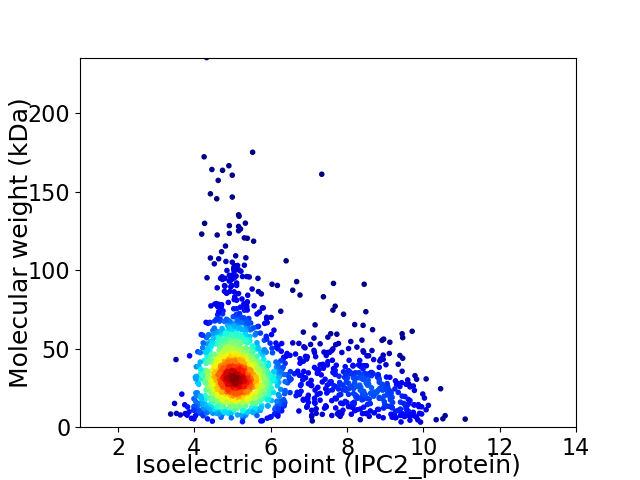
Virtual 2D-PAGE plot for 1739 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1QYB0|E1QYB0_OLSUV ABC transporter related protein OS=Olsenella uli (strain ATCC 49627 / DSM 7084 / CIP 109912 / JCM 12494 / NCIMB 702895 / VPI D76D-27C) OX=633147 GN=Olsu_0248 PE=4 SV=1
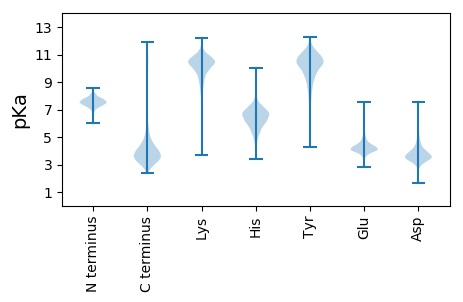
MM1 pKa = 7.17TRR3 pKa = 11.84RR4 pKa = 11.84GLLAASLALAGGALGLAGCGLLRR27 pKa = 11.84GRR29 pKa = 11.84EE30 pKa = 4.05EE31 pKa = 4.13EE32 pKa = 4.36VPDD35 pKa = 3.55VTISSGEE42 pKa = 3.96DD43 pKa = 2.86ALAYY47 pKa = 10.06ALQALGEE54 pKa = 4.24RR55 pKa = 11.84YY56 pKa = 10.31GEE58 pKa = 4.26GFSQAEE64 pKa = 4.23GTEE67 pKa = 4.13ITSYY71 pKa = 10.58MPDD74 pKa = 3.69DD75 pKa = 3.66TDD77 pKa = 4.11LTYY80 pKa = 10.62EE81 pKa = 4.07LCARR85 pKa = 11.84PDD87 pKa = 3.57SDD89 pKa = 3.43PGKK92 pKa = 10.95VFGCDD97 pKa = 2.75VSTDD101 pKa = 3.58RR102 pKa = 11.84EE103 pKa = 4.35TGRR106 pKa = 11.84LLDD109 pKa = 3.95VLEE112 pKa = 5.29DD113 pKa = 5.36DD114 pKa = 3.74YY115 pKa = 11.75TQYY118 pKa = 11.49LFKK121 pKa = 10.99DD122 pKa = 3.48QMEE125 pKa = 4.85GPFLEE130 pKa = 4.67VVRR133 pKa = 11.84GLSGLAGYY141 pKa = 8.66SVQMAEE147 pKa = 4.68PYY149 pKa = 10.76VGDD152 pKa = 3.82RR153 pKa = 11.84DD154 pKa = 3.68WRR156 pKa = 11.84PDD158 pKa = 3.44EE159 pKa = 4.52LDD161 pKa = 3.72AYY163 pKa = 9.9VNNGFTHH170 pKa = 6.26PRR172 pKa = 11.84VDD174 pKa = 3.55VTLMMPADD182 pKa = 3.75GTEE185 pKa = 4.26GEE187 pKa = 4.4WAGTIHH193 pKa = 6.81EE194 pKa = 4.63CLEE197 pKa = 4.78GIWGLGQRR205 pKa = 11.84MYY207 pKa = 11.15VYY209 pKa = 10.77AGLEE213 pKa = 4.26GYY215 pKa = 10.21DD216 pKa = 3.69PYY218 pKa = 11.33SAQLYY223 pKa = 10.36ILDD226 pKa = 3.75STQNEE231 pKa = 4.12VRR233 pKa = 11.84GRR235 pKa = 11.84DD236 pKa = 3.79PEE238 pKa = 4.34PPSVDD243 pKa = 4.12RR244 pKa = 11.84ILEE247 pKa = 4.18LMWPDD252 pKa = 3.27VVYY255 pKa = 10.8RR256 pKa = 11.84DD257 pKa = 5.25GSEE260 pKa = 3.95TWDD263 pKa = 3.56GTGDD267 pKa = 3.68AGDD270 pKa = 3.98PGYY273 pKa = 11.11VKK275 pKa = 10.42HH276 pKa = 6.61PQITWYY282 pKa = 10.03DD283 pKa = 3.48GHH285 pKa = 7.71PVMPP289 pKa = 5.45
MM1 pKa = 7.17TRR3 pKa = 11.84RR4 pKa = 11.84GLLAASLALAGGALGLAGCGLLRR27 pKa = 11.84GRR29 pKa = 11.84EE30 pKa = 4.05EE31 pKa = 4.13EE32 pKa = 4.36VPDD35 pKa = 3.55VTISSGEE42 pKa = 3.96DD43 pKa = 2.86ALAYY47 pKa = 10.06ALQALGEE54 pKa = 4.24RR55 pKa = 11.84YY56 pKa = 10.31GEE58 pKa = 4.26GFSQAEE64 pKa = 4.23GTEE67 pKa = 4.13ITSYY71 pKa = 10.58MPDD74 pKa = 3.69DD75 pKa = 3.66TDD77 pKa = 4.11LTYY80 pKa = 10.62EE81 pKa = 4.07LCARR85 pKa = 11.84PDD87 pKa = 3.57SDD89 pKa = 3.43PGKK92 pKa = 10.95VFGCDD97 pKa = 2.75VSTDD101 pKa = 3.58RR102 pKa = 11.84EE103 pKa = 4.35TGRR106 pKa = 11.84LLDD109 pKa = 3.95VLEE112 pKa = 5.29DD113 pKa = 5.36DD114 pKa = 3.74YY115 pKa = 11.75TQYY118 pKa = 11.49LFKK121 pKa = 10.99DD122 pKa = 3.48QMEE125 pKa = 4.85GPFLEE130 pKa = 4.67VVRR133 pKa = 11.84GLSGLAGYY141 pKa = 8.66SVQMAEE147 pKa = 4.68PYY149 pKa = 10.76VGDD152 pKa = 3.82RR153 pKa = 11.84DD154 pKa = 3.68WRR156 pKa = 11.84PDD158 pKa = 3.44EE159 pKa = 4.52LDD161 pKa = 3.72AYY163 pKa = 9.9VNNGFTHH170 pKa = 6.26PRR172 pKa = 11.84VDD174 pKa = 3.55VTLMMPADD182 pKa = 3.75GTEE185 pKa = 4.26GEE187 pKa = 4.4WAGTIHH193 pKa = 6.81EE194 pKa = 4.63CLEE197 pKa = 4.78GIWGLGQRR205 pKa = 11.84MYY207 pKa = 11.15VYY209 pKa = 10.77AGLEE213 pKa = 4.26GYY215 pKa = 10.21DD216 pKa = 3.69PYY218 pKa = 11.33SAQLYY223 pKa = 10.36ILDD226 pKa = 3.75STQNEE231 pKa = 4.12VRR233 pKa = 11.84GRR235 pKa = 11.84DD236 pKa = 3.79PEE238 pKa = 4.34PPSVDD243 pKa = 4.12RR244 pKa = 11.84ILEE247 pKa = 4.18LMWPDD252 pKa = 3.27VVYY255 pKa = 10.8RR256 pKa = 11.84DD257 pKa = 5.25GSEE260 pKa = 3.95TWDD263 pKa = 3.56GTGDD267 pKa = 3.68AGDD270 pKa = 3.98PGYY273 pKa = 11.11VKK275 pKa = 10.42HH276 pKa = 6.61PQITWYY282 pKa = 10.03DD283 pKa = 3.48GHH285 pKa = 7.71PVMPP289 pKa = 5.45
Molecular weight: 31.79 kDa
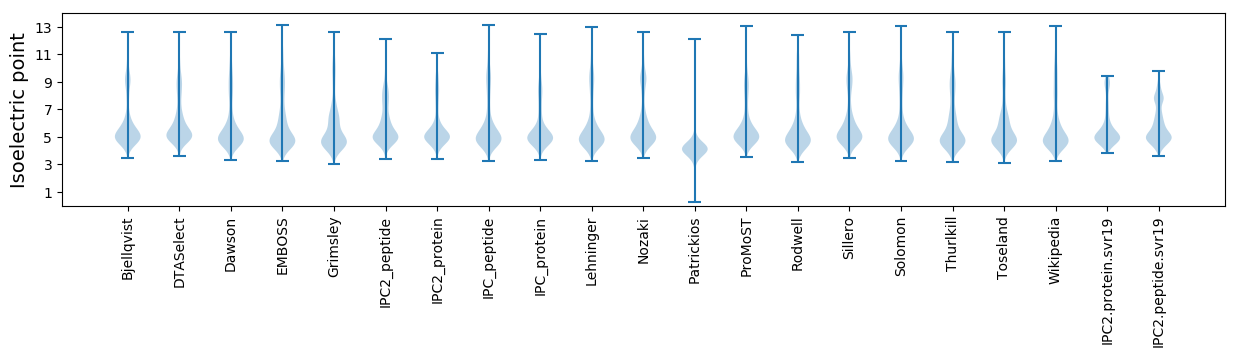
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1QXN3|E1QXN3_OLSUV Putative PTS IIA-like nitrogen-regulatory protein PtsN OS=Olsenella uli (strain ATCC 49627 / DSM 7084 / CIP 109912 / JCM 12494 / NCIMB 702895 / VPI D76D-27C) OX=633147 GN=Olsu_0012 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.62RR10 pKa = 11.84KK11 pKa = 9.56RR12 pKa = 11.84AKK14 pKa = 8.76THH16 pKa = 5.23GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84KK40 pKa = 8.12QLTVV44 pKa = 2.97
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.62RR10 pKa = 11.84KK11 pKa = 9.56RR12 pKa = 11.84AKK14 pKa = 8.76THH16 pKa = 5.23GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84KK40 pKa = 8.12QLTVV44 pKa = 2.97
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
592283 |
31 |
2342 |
340.6 |
36.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.203 ± 0.078 | 1.53 ± 0.026 |
6.552 ± 0.046 | 6.245 ± 0.052 |
3.359 ± 0.033 | 9.006 ± 0.064 |
1.985 ± 0.022 | 4.572 ± 0.053 |
3.047 ± 0.049 | 9.431 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.027 | 2.479 ± 0.031 |
4.262 ± 0.039 | 2.755 ± 0.032 |
6.959 ± 0.063 | 6.292 ± 0.045 |
5.327 ± 0.048 | 8.569 ± 0.051 |
1.066 ± 0.02 | 2.73 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
