
Aquiflexum balticum DSM 16537
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Aquiflexum; Aquiflexum balticum
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
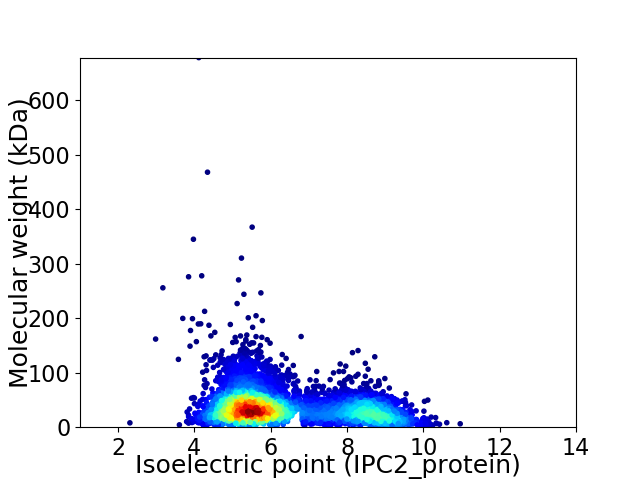
Virtual 2D-PAGE plot for 4889 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
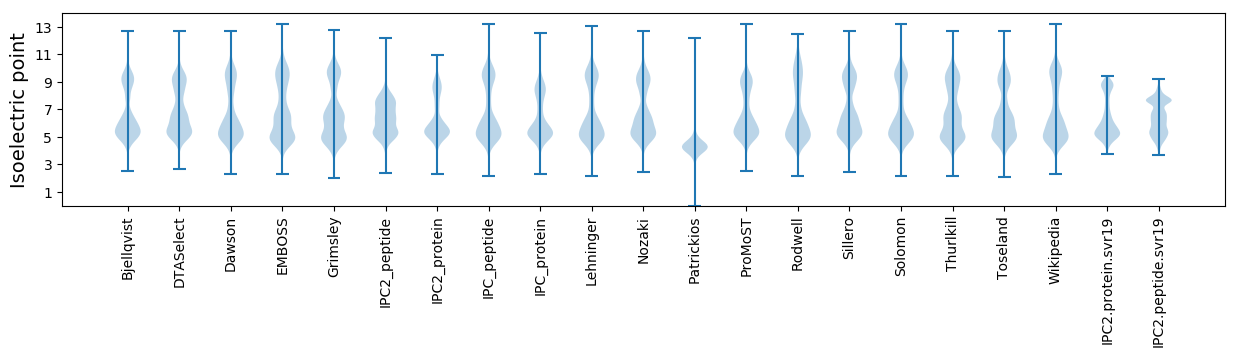
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W2H4H1|A0A1W2H4H1_9BACT Isocitrate dehydrogenase (NADP(+)) OS=Aquiflexum balticum DSM 16537 OX=758820 GN=SAMN00777080_2398 PE=3 SV=1
MM1 pKa = 7.28LALVAFMVACSQFEE15 pKa = 4.14AQGPDD20 pKa = 3.58DD21 pKa = 4.42VNLEE25 pKa = 4.09NAEE28 pKa = 4.11EE29 pKa = 3.93LASAFSMDD37 pKa = 3.78PYY39 pKa = 11.62GLGLEE44 pKa = 4.35NARR47 pKa = 11.84VPLEE51 pKa = 4.29GKK53 pKa = 8.56GTASEE58 pKa = 4.16PVSDD62 pKa = 5.0GGITPYY68 pKa = 10.38IITGTSGGGNRR79 pKa = 11.84TCDD82 pKa = 3.59EE83 pKa = 3.96VATAYY88 pKa = 10.61GIGSFEE94 pKa = 4.63NSFEE98 pKa = 4.32QINTPFTNNTATFGLLTATTDD119 pKa = 3.39GTFVSWSIEE128 pKa = 4.01VPEE131 pKa = 5.35GYY133 pKa = 9.51CVKK136 pKa = 10.64YY137 pKa = 10.18VAAVVKK143 pKa = 10.42GGNDD147 pKa = 3.22ANVYY151 pKa = 9.94FYY153 pKa = 10.86EE154 pKa = 5.91GDD156 pKa = 3.39VSGDD160 pKa = 3.29SGLASPGAGGSGGAASLSNLRR181 pKa = 11.84FCYY184 pKa = 9.85TLEE187 pKa = 4.36EE188 pKa = 4.73KK189 pKa = 10.43PDD191 pKa = 3.66APIVEE196 pKa = 4.37GDD198 pKa = 3.71EE199 pKa = 4.23EE200 pKa = 5.5CFVEE204 pKa = 5.25DD205 pKa = 3.6LVLRR209 pKa = 11.84ATLADD214 pKa = 3.37GVTIPEE220 pKa = 5.14GYY222 pKa = 8.07TLQWYY227 pKa = 9.83KK228 pKa = 10.77KK229 pKa = 9.55VNGEE233 pKa = 4.02YY234 pKa = 9.96VALGEE239 pKa = 4.3GEE241 pKa = 4.52EE242 pKa = 4.72PILDD246 pKa = 3.71EE247 pKa = 4.75VGSIDD252 pKa = 3.88YY253 pKa = 10.5YY254 pKa = 10.72AAFVNGCSSEE264 pKa = 4.01MSNAATLTINPLPGPPTGGDD284 pKa = 3.15QDD286 pKa = 5.09EE287 pKa = 4.59EE288 pKa = 4.57VCDD291 pKa = 4.9FSKK294 pKa = 10.05LTATVKK300 pKa = 10.46DD301 pKa = 3.89VPDD304 pKa = 3.94GVSIVWYY311 pKa = 9.27DD312 pKa = 3.27APTGGNVVTDD322 pKa = 4.15PSLVYY327 pKa = 10.42DD328 pKa = 4.0PSGDD332 pKa = 3.63EE333 pKa = 4.05VQTATFYY340 pKa = 11.62AEE342 pKa = 4.06AVNDD346 pKa = 3.51EE347 pKa = 4.65TEE349 pKa = 4.47CVSATRR355 pKa = 11.84TMVTLTLRR363 pKa = 11.84PCTRR367 pKa = 11.84EE368 pKa = 4.03CYY370 pKa = 10.15DD371 pKa = 4.37DD372 pKa = 4.48EE373 pKa = 4.78SATGRR378 pKa = 11.84GNAFTNRR385 pKa = 11.84RR386 pKa = 11.84PATWFQWNTRR396 pKa = 11.84GEE398 pKa = 4.8LITGVDD404 pKa = 3.68LVYY407 pKa = 10.88GRR409 pKa = 11.84QLEE412 pKa = 4.44KK413 pKa = 10.8AGSVTISGPDD423 pKa = 3.04GGFRR427 pKa = 11.84TITVTLDD434 pKa = 2.98PGFRR438 pKa = 11.84FAQDD442 pKa = 2.82ADD444 pKa = 3.84GNIVSGALKK453 pKa = 10.55VFASNTQFTQFSGFSGALQSNTATIRR479 pKa = 11.84VPDD482 pKa = 3.3TGFYY486 pKa = 10.05FVHH489 pKa = 7.02GDD491 pKa = 3.25IEE493 pKa = 4.54RR494 pKa = 11.84KK495 pKa = 8.89IDD497 pKa = 4.38CPIADD502 pKa = 3.47
MM1 pKa = 7.28LALVAFMVACSQFEE15 pKa = 4.14AQGPDD20 pKa = 3.58DD21 pKa = 4.42VNLEE25 pKa = 4.09NAEE28 pKa = 4.11EE29 pKa = 3.93LASAFSMDD37 pKa = 3.78PYY39 pKa = 11.62GLGLEE44 pKa = 4.35NARR47 pKa = 11.84VPLEE51 pKa = 4.29GKK53 pKa = 8.56GTASEE58 pKa = 4.16PVSDD62 pKa = 5.0GGITPYY68 pKa = 10.38IITGTSGGGNRR79 pKa = 11.84TCDD82 pKa = 3.59EE83 pKa = 3.96VATAYY88 pKa = 10.61GIGSFEE94 pKa = 4.63NSFEE98 pKa = 4.32QINTPFTNNTATFGLLTATTDD119 pKa = 3.39GTFVSWSIEE128 pKa = 4.01VPEE131 pKa = 5.35GYY133 pKa = 9.51CVKK136 pKa = 10.64YY137 pKa = 10.18VAAVVKK143 pKa = 10.42GGNDD147 pKa = 3.22ANVYY151 pKa = 9.94FYY153 pKa = 10.86EE154 pKa = 5.91GDD156 pKa = 3.39VSGDD160 pKa = 3.29SGLASPGAGGSGGAASLSNLRR181 pKa = 11.84FCYY184 pKa = 9.85TLEE187 pKa = 4.36EE188 pKa = 4.73KK189 pKa = 10.43PDD191 pKa = 3.66APIVEE196 pKa = 4.37GDD198 pKa = 3.71EE199 pKa = 4.23EE200 pKa = 5.5CFVEE204 pKa = 5.25DD205 pKa = 3.6LVLRR209 pKa = 11.84ATLADD214 pKa = 3.37GVTIPEE220 pKa = 5.14GYY222 pKa = 8.07TLQWYY227 pKa = 9.83KK228 pKa = 10.77KK229 pKa = 9.55VNGEE233 pKa = 4.02YY234 pKa = 9.96VALGEE239 pKa = 4.3GEE241 pKa = 4.52EE242 pKa = 4.72PILDD246 pKa = 3.71EE247 pKa = 4.75VGSIDD252 pKa = 3.88YY253 pKa = 10.5YY254 pKa = 10.72AAFVNGCSSEE264 pKa = 4.01MSNAATLTINPLPGPPTGGDD284 pKa = 3.15QDD286 pKa = 5.09EE287 pKa = 4.59EE288 pKa = 4.57VCDD291 pKa = 4.9FSKK294 pKa = 10.05LTATVKK300 pKa = 10.46DD301 pKa = 3.89VPDD304 pKa = 3.94GVSIVWYY311 pKa = 9.27DD312 pKa = 3.27APTGGNVVTDD322 pKa = 4.15PSLVYY327 pKa = 10.42DD328 pKa = 4.0PSGDD332 pKa = 3.63EE333 pKa = 4.05VQTATFYY340 pKa = 11.62AEE342 pKa = 4.06AVNDD346 pKa = 3.51EE347 pKa = 4.65TEE349 pKa = 4.47CVSATRR355 pKa = 11.84TMVTLTLRR363 pKa = 11.84PCTRR367 pKa = 11.84EE368 pKa = 4.03CYY370 pKa = 10.15DD371 pKa = 4.37DD372 pKa = 4.48EE373 pKa = 4.78SATGRR378 pKa = 11.84GNAFTNRR385 pKa = 11.84RR386 pKa = 11.84PATWFQWNTRR396 pKa = 11.84GEE398 pKa = 4.8LITGVDD404 pKa = 3.68LVYY407 pKa = 10.88GRR409 pKa = 11.84QLEE412 pKa = 4.44KK413 pKa = 10.8AGSVTISGPDD423 pKa = 3.04GGFRR427 pKa = 11.84TITVTLDD434 pKa = 2.98PGFRR438 pKa = 11.84FAQDD442 pKa = 2.82ADD444 pKa = 3.84GNIVSGALKK453 pKa = 10.55VFASNTQFTQFSGFSGALQSNTATIRR479 pKa = 11.84VPDD482 pKa = 3.3TGFYY486 pKa = 10.05FVHH489 pKa = 7.02GDD491 pKa = 3.25IEE493 pKa = 4.54RR494 pKa = 11.84KK495 pKa = 8.89IDD497 pKa = 4.38CPIADD502 pKa = 3.47
Molecular weight: 53.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W2H4E2|A0A1W2H4E2_9BACT Uncharacterized protein OS=Aquiflexum balticum DSM 16537 OX=758820 GN=SAMN00777080_2427 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.78HH16 pKa = 3.85GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.41SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.19GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.78HH16 pKa = 3.85GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.41SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.19GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1724006 |
29 |
6369 |
352.6 |
39.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.171 ± 0.038 | 0.686 ± 0.011 |
5.405 ± 0.029 | 6.907 ± 0.035 |
5.578 ± 0.028 | 7.045 ± 0.04 |
1.824 ± 0.016 | 7.69 ± 0.031 |
7.046 ± 0.044 | 9.67 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.019 | 5.504 ± 0.036 |
3.938 ± 0.019 | 3.513 ± 0.019 |
3.913 ± 0.024 | 6.583 ± 0.026 |
5.024 ± 0.037 | 6.053 ± 0.028 |
1.288 ± 0.013 | 3.726 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
