
Duck associated cyclovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus
Average proteome isoelectric point is 8.71
Get precalculated fractions of proteins
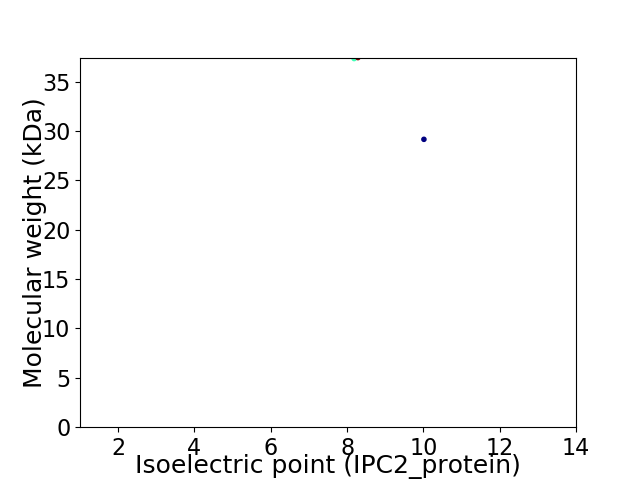
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y0SVU5|A0A1Y0SVU5_9CIRC ATP-dependent helicase Rep OS=Duck associated cyclovirus 1 OX=2006585 PE=3 SV=1
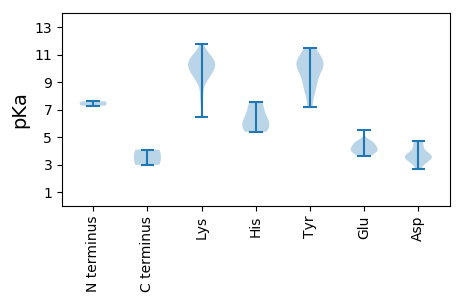
MM1 pKa = 7.28QRR3 pKa = 11.84KK4 pKa = 9.22CYY6 pKa = 10.43SWTLNNYY13 pKa = 7.18TSEE16 pKa = 4.63DD17 pKa = 3.23IEE19 pKa = 4.63RR20 pKa = 11.84IRR22 pKa = 11.84QHH24 pKa = 5.54EE25 pKa = 4.24HH26 pKa = 6.15LFDD29 pKa = 3.56YY30 pKa = 10.48AIWGRR35 pKa = 11.84EE36 pKa = 3.71IAPEE40 pKa = 3.97TGTPHH45 pKa = 5.63LQGYY49 pKa = 8.07FKK51 pKa = 10.98FKK53 pKa = 9.87TKK55 pKa = 10.12QRR57 pKa = 11.84FNTVRR62 pKa = 11.84AILGGRR68 pKa = 11.84GHH70 pKa = 7.53LEE72 pKa = 3.94GSKK75 pKa = 10.81GSPKK79 pKa = 10.08QNHH82 pKa = 7.04DD83 pKa = 3.66YY84 pKa = 10.37CAKK87 pKa = 10.09SGDD90 pKa = 3.92YY91 pKa = 8.45EE92 pKa = 4.57TVGDD96 pKa = 4.74LVIASQSSLEE106 pKa = 4.6LVCQMLKK113 pKa = 10.9DD114 pKa = 4.14GVRR117 pKa = 11.84LAQVAADD124 pKa = 3.77YY125 pKa = 10.21PSIFVRR131 pKa = 11.84HH132 pKa = 5.38HH133 pKa = 6.08RR134 pKa = 11.84GLRR137 pKa = 11.84EE138 pKa = 3.65LSLILGVVKK147 pKa = 10.38PRR149 pKa = 11.84DD150 pKa = 3.57FKK152 pKa = 9.43TGKK155 pKa = 8.02WAAARR160 pKa = 11.84SIPEE164 pKa = 3.72SLSRR168 pKa = 11.84FRR170 pKa = 11.84ANLLLTVRR178 pKa = 11.84WMILSLLVLVSEE190 pKa = 4.4VLVFYY195 pKa = 10.33GASGTGKK202 pKa = 9.79SRR204 pKa = 11.84RR205 pKa = 11.84AAEE208 pKa = 4.12LCGSEE213 pKa = 3.79STYY216 pKa = 10.88YY217 pKa = 10.48KK218 pKa = 10.5PRR220 pKa = 11.84GKK222 pKa = 9.0WWDD225 pKa = 3.58GYY227 pKa = 11.49ANQDD231 pKa = 3.1NVIIDD236 pKa = 4.57DD237 pKa = 4.55FYY239 pKa = 11.37GWLSFDD245 pKa = 3.4EE246 pKa = 4.63FLRR249 pKa = 11.84IADD252 pKa = 4.55RR253 pKa = 11.84YY254 pKa = 7.36PCRR257 pKa = 11.84VEE259 pKa = 4.22VKK261 pKa = 10.58GGFAEE266 pKa = 4.7FTSKK270 pKa = 10.8RR271 pKa = 11.84IVITSNQAPSQWWRR285 pKa = 11.84GEE287 pKa = 4.01WFKK290 pKa = 11.79GEE292 pKa = 3.85QLKK295 pKa = 11.05ALTRR299 pKa = 11.84RR300 pKa = 11.84LDD302 pKa = 3.54VVCFYY307 pKa = 10.56SAHH310 pKa = 6.41SGLPHH315 pKa = 7.56EE316 pKa = 5.5INCQGDD322 pKa = 3.83CDD324 pKa = 4.29KK325 pKa = 11.23EE326 pKa = 4.08
MM1 pKa = 7.28QRR3 pKa = 11.84KK4 pKa = 9.22CYY6 pKa = 10.43SWTLNNYY13 pKa = 7.18TSEE16 pKa = 4.63DD17 pKa = 3.23IEE19 pKa = 4.63RR20 pKa = 11.84IRR22 pKa = 11.84QHH24 pKa = 5.54EE25 pKa = 4.24HH26 pKa = 6.15LFDD29 pKa = 3.56YY30 pKa = 10.48AIWGRR35 pKa = 11.84EE36 pKa = 3.71IAPEE40 pKa = 3.97TGTPHH45 pKa = 5.63LQGYY49 pKa = 8.07FKK51 pKa = 10.98FKK53 pKa = 9.87TKK55 pKa = 10.12QRR57 pKa = 11.84FNTVRR62 pKa = 11.84AILGGRR68 pKa = 11.84GHH70 pKa = 7.53LEE72 pKa = 3.94GSKK75 pKa = 10.81GSPKK79 pKa = 10.08QNHH82 pKa = 7.04DD83 pKa = 3.66YY84 pKa = 10.37CAKK87 pKa = 10.09SGDD90 pKa = 3.92YY91 pKa = 8.45EE92 pKa = 4.57TVGDD96 pKa = 4.74LVIASQSSLEE106 pKa = 4.6LVCQMLKK113 pKa = 10.9DD114 pKa = 4.14GVRR117 pKa = 11.84LAQVAADD124 pKa = 3.77YY125 pKa = 10.21PSIFVRR131 pKa = 11.84HH132 pKa = 5.38HH133 pKa = 6.08RR134 pKa = 11.84GLRR137 pKa = 11.84EE138 pKa = 3.65LSLILGVVKK147 pKa = 10.38PRR149 pKa = 11.84DD150 pKa = 3.57FKK152 pKa = 9.43TGKK155 pKa = 8.02WAAARR160 pKa = 11.84SIPEE164 pKa = 3.72SLSRR168 pKa = 11.84FRR170 pKa = 11.84ANLLLTVRR178 pKa = 11.84WMILSLLVLVSEE190 pKa = 4.4VLVFYY195 pKa = 10.33GASGTGKK202 pKa = 9.79SRR204 pKa = 11.84RR205 pKa = 11.84AAEE208 pKa = 4.12LCGSEE213 pKa = 3.79STYY216 pKa = 10.88YY217 pKa = 10.48KK218 pKa = 10.5PRR220 pKa = 11.84GKK222 pKa = 9.0WWDD225 pKa = 3.58GYY227 pKa = 11.49ANQDD231 pKa = 3.1NVIIDD236 pKa = 4.57DD237 pKa = 4.55FYY239 pKa = 11.37GWLSFDD245 pKa = 3.4EE246 pKa = 4.63FLRR249 pKa = 11.84IADD252 pKa = 4.55RR253 pKa = 11.84YY254 pKa = 7.36PCRR257 pKa = 11.84VEE259 pKa = 4.22VKK261 pKa = 10.58GGFAEE266 pKa = 4.7FTSKK270 pKa = 10.8RR271 pKa = 11.84IVITSNQAPSQWWRR285 pKa = 11.84GEE287 pKa = 4.01WFKK290 pKa = 11.79GEE292 pKa = 3.85QLKK295 pKa = 11.05ALTRR299 pKa = 11.84RR300 pKa = 11.84LDD302 pKa = 3.54VVCFYY307 pKa = 10.56SAHH310 pKa = 6.41SGLPHH315 pKa = 7.56EE316 pKa = 5.5INCQGDD322 pKa = 3.83CDD324 pKa = 4.29KK325 pKa = 11.23EE326 pKa = 4.08
Molecular weight: 37.34 kDa
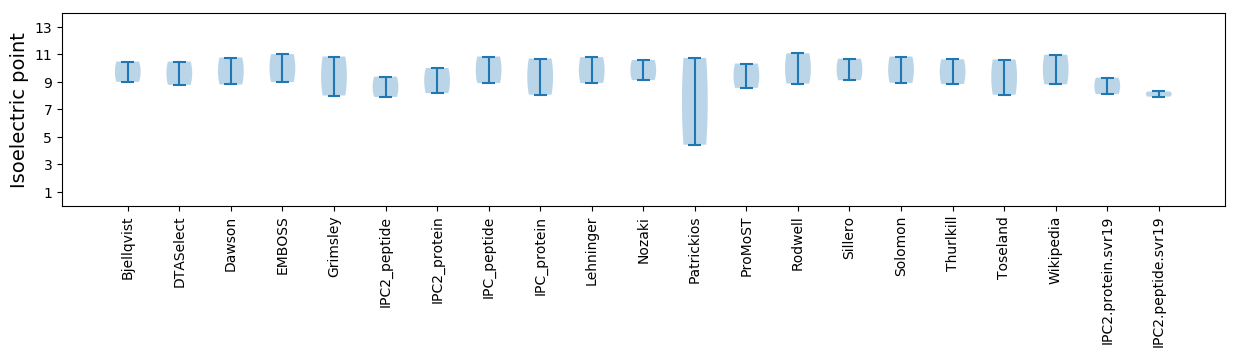
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y0SVU5|A0A1Y0SVU5_9CIRC ATP-dependent helicase Rep OS=Duck associated cyclovirus 1 OX=2006585 PE=3 SV=1
MM1 pKa = 7.59ANYY4 pKa = 9.47RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.8KK8 pKa = 10.13KK9 pKa = 9.61YY10 pKa = 8.71GRR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 8.67RR15 pKa = 11.84PRR17 pKa = 11.84PLRR20 pKa = 11.84VRR22 pKa = 11.84LYY24 pKa = 8.91NPKK27 pKa = 10.03RR28 pKa = 11.84NRR30 pKa = 11.84LRR32 pKa = 11.84RR33 pKa = 11.84TQRR36 pKa = 11.84RR37 pKa = 11.84LKK39 pKa = 9.22NDD41 pKa = 2.74KK42 pKa = 10.26YY43 pKa = 9.93FTRR46 pKa = 11.84FMSWTSVDD54 pKa = 3.39VDD56 pKa = 3.73PKK58 pKa = 10.62IGQGWYY64 pKa = 9.78LAPNYY69 pKa = 9.84KK70 pKa = 10.41SNPGFLQLAEE80 pKa = 4.24QYY82 pKa = 11.09SEE84 pKa = 4.22FKK86 pKa = 9.2IHH88 pKa = 6.58KK89 pKa = 7.81VAARR93 pKa = 11.84ITPGLNEE100 pKa = 4.33TYY102 pKa = 10.43LAEE105 pKa = 4.05QGLRR109 pKa = 11.84YY110 pKa = 9.65AYY112 pKa = 10.33CPWQQDD118 pKa = 3.44NGGSPSTNDD127 pKa = 2.71IVSNGTDD134 pKa = 3.28LTYY137 pKa = 11.02GKK139 pKa = 10.62LMNVPWAKK147 pKa = 10.12SKK149 pKa = 10.61RR150 pKa = 11.84LHH152 pKa = 5.58QSATIVYY159 pKa = 9.61KK160 pKa = 9.69PKK162 pKa = 10.36CNQAIAFAPRR172 pKa = 11.84TSSGANGNQRR182 pKa = 11.84VISFPWVTTQAMSMGALPGMSGIMLAFDD210 pKa = 3.5QQDD213 pKa = 3.73PLSVIGKK220 pKa = 6.49PLRR223 pKa = 11.84VQVEE227 pKa = 4.05FFLYY231 pKa = 8.66VTFRR235 pKa = 11.84KK236 pKa = 9.59RR237 pKa = 11.84RR238 pKa = 11.84NPMLQTPTPFVMDD251 pKa = 3.16QQ252 pKa = 2.98
MM1 pKa = 7.59ANYY4 pKa = 9.47RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.8KK8 pKa = 10.13KK9 pKa = 9.61YY10 pKa = 8.71GRR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 8.67RR15 pKa = 11.84PRR17 pKa = 11.84PLRR20 pKa = 11.84VRR22 pKa = 11.84LYY24 pKa = 8.91NPKK27 pKa = 10.03RR28 pKa = 11.84NRR30 pKa = 11.84LRR32 pKa = 11.84RR33 pKa = 11.84TQRR36 pKa = 11.84RR37 pKa = 11.84LKK39 pKa = 9.22NDD41 pKa = 2.74KK42 pKa = 10.26YY43 pKa = 9.93FTRR46 pKa = 11.84FMSWTSVDD54 pKa = 3.39VDD56 pKa = 3.73PKK58 pKa = 10.62IGQGWYY64 pKa = 9.78LAPNYY69 pKa = 9.84KK70 pKa = 10.41SNPGFLQLAEE80 pKa = 4.24QYY82 pKa = 11.09SEE84 pKa = 4.22FKK86 pKa = 9.2IHH88 pKa = 6.58KK89 pKa = 7.81VAARR93 pKa = 11.84ITPGLNEE100 pKa = 4.33TYY102 pKa = 10.43LAEE105 pKa = 4.05QGLRR109 pKa = 11.84YY110 pKa = 9.65AYY112 pKa = 10.33CPWQQDD118 pKa = 3.44NGGSPSTNDD127 pKa = 2.71IVSNGTDD134 pKa = 3.28LTYY137 pKa = 11.02GKK139 pKa = 10.62LMNVPWAKK147 pKa = 10.12SKK149 pKa = 10.61RR150 pKa = 11.84LHH152 pKa = 5.58QSATIVYY159 pKa = 9.61KK160 pKa = 9.69PKK162 pKa = 10.36CNQAIAFAPRR172 pKa = 11.84TSSGANGNQRR182 pKa = 11.84VISFPWVTTQAMSMGALPGMSGIMLAFDD210 pKa = 3.5QQDD213 pKa = 3.73PLSVIGKK220 pKa = 6.49PLRR223 pKa = 11.84VQVEE227 pKa = 4.05FFLYY231 pKa = 8.66VTFRR235 pKa = 11.84KK236 pKa = 9.59RR237 pKa = 11.84RR238 pKa = 11.84NPMLQTPTPFVMDD251 pKa = 3.16QQ252 pKa = 2.98
Molecular weight: 29.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
578 |
252 |
326 |
289.0 |
33.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.401 ± 0.033 | 1.73 ± 0.584 |
4.498 ± 0.578 | 4.325 ± 1.46 |
4.498 ± 0.083 | 7.439 ± 0.68 |
1.903 ± 0.692 | 4.325 ± 0.47 |
6.228 ± 0.323 | 8.478 ± 0.585 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.076 ± 0.932 | 4.152 ± 1.122 |
5.017 ± 1.573 | 5.017 ± 0.83 |
8.651 ± 0.545 | 6.92 ± 0.604 |
5.017 ± 0.583 | 5.882 ± 0.204 |
2.595 ± 0.381 | 4.844 ± 0.443 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
