
Tomelloso virus
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Nudiviridae; Alphanudivirus; Drosophila melanogaster nudivirus B
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
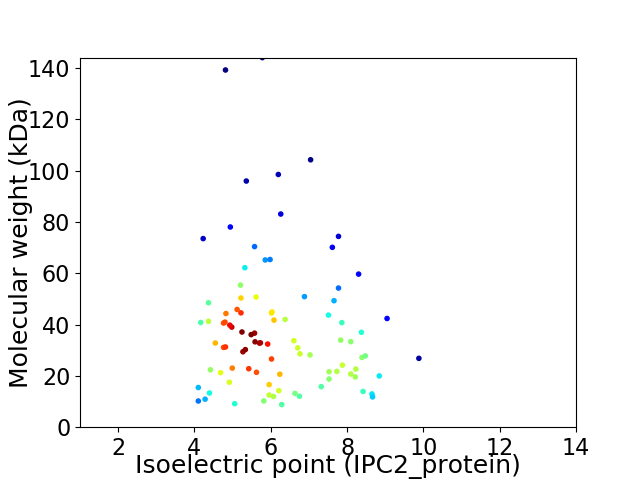
Virtual 2D-PAGE plot for 93 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
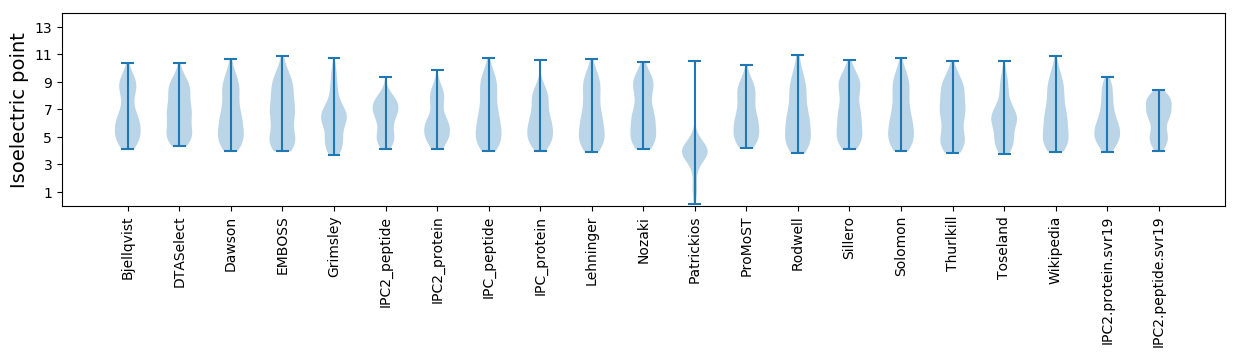
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4T2W2|A0A2H4T2W2_9VIRU OrNV gp035-like protein OS=Tomelloso virus OX=2053981 PE=4 SV=1
MM1 pKa = 7.56DD2 pKa = 4.78QIHH5 pKa = 6.93DD6 pKa = 4.09AFVEE10 pKa = 4.05LLTLGIITDD19 pKa = 3.66SDD21 pKa = 4.01EE22 pKa = 4.46EE23 pKa = 4.3NVTIDD28 pKa = 5.01LEE30 pKa = 4.29ALYY33 pKa = 10.89SLGYY37 pKa = 9.58VLVYY41 pKa = 10.96NLTIPVFTTILNYY54 pKa = 10.77NYY56 pKa = 9.28MYY58 pKa = 10.92DD59 pKa = 4.26GILYY63 pKa = 8.34TSNVVPRR70 pKa = 11.84RR71 pKa = 11.84ALSTFVLSKK80 pKa = 9.99IQTIGVSDD88 pKa = 4.88FKK90 pKa = 11.59
MM1 pKa = 7.56DD2 pKa = 4.78QIHH5 pKa = 6.93DD6 pKa = 4.09AFVEE10 pKa = 4.05LLTLGIITDD19 pKa = 3.66SDD21 pKa = 4.01EE22 pKa = 4.46EE23 pKa = 4.3NVTIDD28 pKa = 5.01LEE30 pKa = 4.29ALYY33 pKa = 10.89SLGYY37 pKa = 9.58VLVYY41 pKa = 10.96NLTIPVFTTILNYY54 pKa = 10.77NYY56 pKa = 9.28MYY58 pKa = 10.92DD59 pKa = 4.26GILYY63 pKa = 8.34TSNVVPRR70 pKa = 11.84RR71 pKa = 11.84ALSTFVLSKK80 pKa = 9.99IQTIGVSDD88 pKa = 4.88FKK90 pKa = 11.59
Molecular weight: 10.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4T2T7|A0A2H4T2T7_9VIRU GrBNV gp61-like protein OS=Tomelloso virus OX=2053981 PE=4 SV=1
MM1 pKa = 7.24NVYY4 pKa = 8.28ITNISEE10 pKa = 4.98FINSIHH16 pKa = 7.71DD17 pKa = 4.36YY18 pKa = 8.33ITEE21 pKa = 4.06RR22 pKa = 11.84YY23 pKa = 9.17EE24 pKa = 3.55IVPRR28 pKa = 11.84DD29 pKa = 3.95FVTKK33 pKa = 10.56LIVGNNALAWHH44 pKa = 6.69SEE46 pKa = 4.14TQQMEE51 pKa = 4.26EE52 pKa = 3.78FHH54 pKa = 6.09CQKK57 pKa = 10.3FKK59 pKa = 11.11KK60 pKa = 9.63HH61 pKa = 5.02LHH63 pKa = 3.98IHH65 pKa = 6.1KK66 pKa = 9.69NYY68 pKa = 10.25KK69 pKa = 10.11NIVYY73 pKa = 9.92CCTEE77 pKa = 3.43VDD79 pKa = 4.41IIALKK84 pKa = 10.2YY85 pKa = 10.51LNASIFGNLKK95 pKa = 10.46NEE97 pKa = 4.78IISSLHH103 pKa = 5.13SWLPAATFQHH113 pKa = 5.19VRR115 pKa = 11.84VTIKK119 pKa = 10.43PATSKK124 pKa = 10.12IDD126 pKa = 3.48SFFEE130 pKa = 3.97PSYY133 pKa = 11.19LFYY136 pKa = 10.49LQSYY140 pKa = 8.66IDD142 pKa = 4.38HH143 pKa = 7.62PIDD146 pKa = 2.85ICYY149 pKa = 9.89TYY151 pKa = 10.22FHH153 pKa = 7.14SNIVFYY159 pKa = 10.9KK160 pKa = 10.01SYY162 pKa = 8.92TLKK165 pKa = 10.78GYY167 pKa = 9.34RR168 pKa = 11.84EE169 pKa = 4.01KK170 pKa = 11.18LIMVRR175 pKa = 11.84ARR177 pKa = 11.84SPSPARR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84SRR189 pKa = 11.84SPSRR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84SPARR199 pKa = 11.84RR200 pKa = 11.84SRR202 pKa = 11.84SSVKK206 pKa = 10.04KK207 pKa = 9.06RR208 pKa = 11.84AGRR211 pKa = 11.84PRR213 pKa = 11.84SRR215 pKa = 11.84SRR217 pKa = 11.84SRR219 pKa = 11.84SRR221 pKa = 11.84SRR223 pKa = 11.84SRR225 pKa = 11.84SS226 pKa = 3.31
MM1 pKa = 7.24NVYY4 pKa = 8.28ITNISEE10 pKa = 4.98FINSIHH16 pKa = 7.71DD17 pKa = 4.36YY18 pKa = 8.33ITEE21 pKa = 4.06RR22 pKa = 11.84YY23 pKa = 9.17EE24 pKa = 3.55IVPRR28 pKa = 11.84DD29 pKa = 3.95FVTKK33 pKa = 10.56LIVGNNALAWHH44 pKa = 6.69SEE46 pKa = 4.14TQQMEE51 pKa = 4.26EE52 pKa = 3.78FHH54 pKa = 6.09CQKK57 pKa = 10.3FKK59 pKa = 11.11KK60 pKa = 9.63HH61 pKa = 5.02LHH63 pKa = 3.98IHH65 pKa = 6.1KK66 pKa = 9.69NYY68 pKa = 10.25KK69 pKa = 10.11NIVYY73 pKa = 9.92CCTEE77 pKa = 3.43VDD79 pKa = 4.41IIALKK84 pKa = 10.2YY85 pKa = 10.51LNASIFGNLKK95 pKa = 10.46NEE97 pKa = 4.78IISSLHH103 pKa = 5.13SWLPAATFQHH113 pKa = 5.19VRR115 pKa = 11.84VTIKK119 pKa = 10.43PATSKK124 pKa = 10.12IDD126 pKa = 3.48SFFEE130 pKa = 3.97PSYY133 pKa = 11.19LFYY136 pKa = 10.49LQSYY140 pKa = 8.66IDD142 pKa = 4.38HH143 pKa = 7.62PIDD146 pKa = 2.85ICYY149 pKa = 9.89TYY151 pKa = 10.22FHH153 pKa = 7.14SNIVFYY159 pKa = 10.9KK160 pKa = 10.01SYY162 pKa = 8.92TLKK165 pKa = 10.78GYY167 pKa = 9.34RR168 pKa = 11.84EE169 pKa = 4.01KK170 pKa = 11.18LIMVRR175 pKa = 11.84ARR177 pKa = 11.84SPSPARR183 pKa = 11.84RR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84SRR189 pKa = 11.84SPSRR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84SPARR199 pKa = 11.84RR200 pKa = 11.84SRR202 pKa = 11.84SSVKK206 pKa = 10.04KK207 pKa = 9.06RR208 pKa = 11.84AGRR211 pKa = 11.84PRR213 pKa = 11.84SRR215 pKa = 11.84SRR217 pKa = 11.84SRR219 pKa = 11.84SRR221 pKa = 11.84SRR223 pKa = 11.84SRR225 pKa = 11.84SS226 pKa = 3.31
Molecular weight: 26.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
31393 |
78 |
1252 |
337.6 |
38.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.326 ± 0.165 | 2.16 ± 0.115 |
6.342 ± 0.184 | 5.562 ± 0.183 |
4.405 ± 0.164 | 3.692 ± 0.116 |
1.899 ± 0.11 | 8.04 ± 0.134 |
6.619 ± 0.193 | 9.002 ± 0.165 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.631 ± 0.101 | 6.699 ± 0.159 |
4.237 ± 0.152 | 3.504 ± 0.158 |
4.154 ± 0.169 | 8.113 ± 0.345 |
6.384 ± 0.19 | 5.68 ± 0.158 |
0.714 ± 0.053 | 4.839 ± 0.155 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
