
Bemisia-associated genomovirus NfO
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus bemta1
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
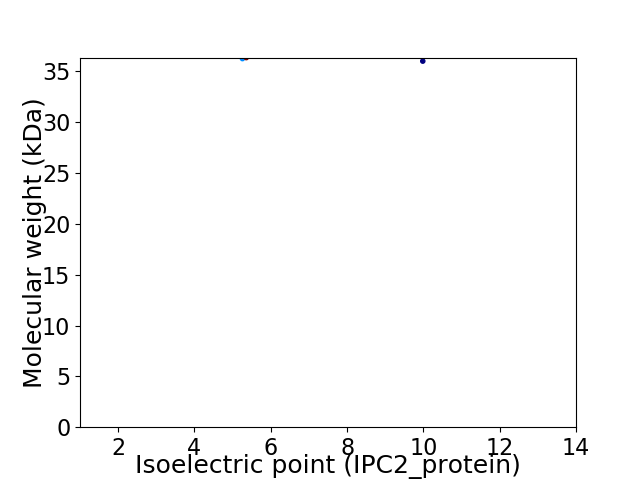
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2X293|A0A1Z2X293_9VIRU Spliced replication-associated protein OS=Bemisia-associated genomovirus NfO OX=1986479 GN=Rep PE=3 SV=2
MM1 pKa = 8.01PFRR4 pKa = 11.84FQAKK8 pKa = 9.39YY9 pKa = 11.04GLLTYY14 pKa = 7.2PQCGDD19 pKa = 3.73LDD21 pKa = 3.59PWHH24 pKa = 6.69IVDD27 pKa = 4.21MLGDD31 pKa = 4.37LGAEE35 pKa = 4.22CIIGRR40 pKa = 11.84EE41 pKa = 4.03DD42 pKa = 3.19HH43 pKa = 6.47VAGGVHH49 pKa = 5.93LHH51 pKa = 6.28AFFMFEE57 pKa = 4.26RR58 pKa = 11.84KK59 pKa = 9.22FEE61 pKa = 4.24SRR63 pKa = 11.84NVRR66 pKa = 11.84VFDD69 pKa = 4.3VDD71 pKa = 3.32GHH73 pKa = 6.37HH74 pKa = 7.12PNVVPGYY81 pKa = 7.34STPEE85 pKa = 4.02KK86 pKa = 9.74GWTYY90 pKa = 9.57ATKK93 pKa = 10.78DD94 pKa = 3.22GDD96 pKa = 3.74IVAGGLEE103 pKa = 4.36CPRR106 pKa = 11.84MRR108 pKa = 11.84EE109 pKa = 4.13EE110 pKa = 4.1VSGASEE116 pKa = 3.64KK117 pKa = 9.97WSRR120 pKa = 11.84ILLSEE125 pKa = 4.03SRR127 pKa = 11.84DD128 pKa = 3.58EE129 pKa = 4.44FFEE132 pKa = 4.41TVARR136 pKa = 11.84LDD138 pKa = 3.64PRR140 pKa = 11.84ALCINFGSLRR150 pKa = 11.84AYY152 pKa = 10.41ADD154 pKa = 2.68WRR156 pKa = 11.84YY157 pKa = 10.15RR158 pKa = 11.84PTRR161 pKa = 11.84DD162 pKa = 3.17PYY164 pKa = 9.54VTPEE168 pKa = 4.01GLSFDD173 pKa = 3.45TSGFPEE179 pKa = 4.54LDD181 pKa = 2.82QWVRR185 pKa = 11.84EE186 pKa = 4.29SLVGLPGTRR195 pKa = 11.84PRR197 pKa = 11.84SLIIIGDD204 pKa = 3.64TRR206 pKa = 11.84LGKK209 pKa = 7.79TLWARR214 pKa = 11.84SLGKK218 pKa = 9.39HH219 pKa = 5.95AYY221 pKa = 9.9FGGLFCLDD229 pKa = 3.79EE230 pKa = 4.48SLEE233 pKa = 4.3DD234 pKa = 2.93VDD236 pKa = 5.06YY237 pKa = 11.57AVFDD241 pKa = 4.16DD242 pKa = 4.05MQGGLEE248 pKa = 4.23FFHH251 pKa = 7.36SYY253 pKa = 10.65KK254 pKa = 10.45FWLGAQKK261 pKa = 10.36QFYY264 pKa = 8.95ATDD267 pKa = 3.37KK268 pKa = 10.83YY269 pKa = 10.66RR270 pKa = 11.84GKK272 pKa = 10.87KK273 pKa = 9.35LVDD276 pKa = 2.98WGRR279 pKa = 11.84PSIYY283 pKa = 9.9ISNTNPLADD292 pKa = 4.19KK293 pKa = 10.9GADD296 pKa = 3.53VDD298 pKa = 4.06WLMGNCTIVHH308 pKa = 6.27VDD310 pKa = 3.28TAIFRR315 pKa = 11.84ANTEE319 pKa = 3.84
MM1 pKa = 8.01PFRR4 pKa = 11.84FQAKK8 pKa = 9.39YY9 pKa = 11.04GLLTYY14 pKa = 7.2PQCGDD19 pKa = 3.73LDD21 pKa = 3.59PWHH24 pKa = 6.69IVDD27 pKa = 4.21MLGDD31 pKa = 4.37LGAEE35 pKa = 4.22CIIGRR40 pKa = 11.84EE41 pKa = 4.03DD42 pKa = 3.19HH43 pKa = 6.47VAGGVHH49 pKa = 5.93LHH51 pKa = 6.28AFFMFEE57 pKa = 4.26RR58 pKa = 11.84KK59 pKa = 9.22FEE61 pKa = 4.24SRR63 pKa = 11.84NVRR66 pKa = 11.84VFDD69 pKa = 4.3VDD71 pKa = 3.32GHH73 pKa = 6.37HH74 pKa = 7.12PNVVPGYY81 pKa = 7.34STPEE85 pKa = 4.02KK86 pKa = 9.74GWTYY90 pKa = 9.57ATKK93 pKa = 10.78DD94 pKa = 3.22GDD96 pKa = 3.74IVAGGLEE103 pKa = 4.36CPRR106 pKa = 11.84MRR108 pKa = 11.84EE109 pKa = 4.13EE110 pKa = 4.1VSGASEE116 pKa = 3.64KK117 pKa = 9.97WSRR120 pKa = 11.84ILLSEE125 pKa = 4.03SRR127 pKa = 11.84DD128 pKa = 3.58EE129 pKa = 4.44FFEE132 pKa = 4.41TVARR136 pKa = 11.84LDD138 pKa = 3.64PRR140 pKa = 11.84ALCINFGSLRR150 pKa = 11.84AYY152 pKa = 10.41ADD154 pKa = 2.68WRR156 pKa = 11.84YY157 pKa = 10.15RR158 pKa = 11.84PTRR161 pKa = 11.84DD162 pKa = 3.17PYY164 pKa = 9.54VTPEE168 pKa = 4.01GLSFDD173 pKa = 3.45TSGFPEE179 pKa = 4.54LDD181 pKa = 2.82QWVRR185 pKa = 11.84EE186 pKa = 4.29SLVGLPGTRR195 pKa = 11.84PRR197 pKa = 11.84SLIIIGDD204 pKa = 3.64TRR206 pKa = 11.84LGKK209 pKa = 7.79TLWARR214 pKa = 11.84SLGKK218 pKa = 9.39HH219 pKa = 5.95AYY221 pKa = 9.9FGGLFCLDD229 pKa = 3.79EE230 pKa = 4.48SLEE233 pKa = 4.3DD234 pKa = 2.93VDD236 pKa = 5.06YY237 pKa = 11.57AVFDD241 pKa = 4.16DD242 pKa = 4.05MQGGLEE248 pKa = 4.23FFHH251 pKa = 7.36SYY253 pKa = 10.65KK254 pKa = 10.45FWLGAQKK261 pKa = 10.36QFYY264 pKa = 8.95ATDD267 pKa = 3.37KK268 pKa = 10.83YY269 pKa = 10.66RR270 pKa = 11.84GKK272 pKa = 10.87KK273 pKa = 9.35LVDD276 pKa = 2.98WGRR279 pKa = 11.84PSIYY283 pKa = 9.9ISNTNPLADD292 pKa = 4.19KK293 pKa = 10.9GADD296 pKa = 3.53VDD298 pKa = 4.06WLMGNCTIVHH308 pKa = 6.27VDD310 pKa = 3.28TAIFRR315 pKa = 11.84ANTEE319 pKa = 3.84
Molecular weight: 36.25 kDa
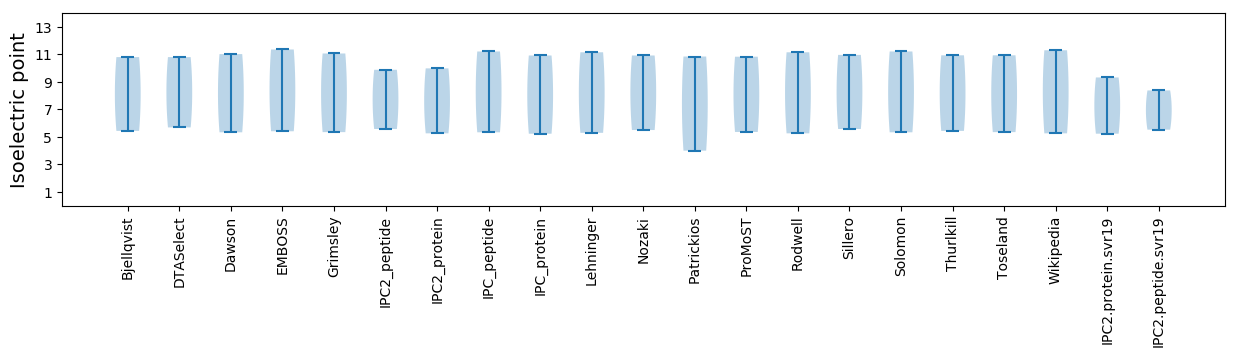
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2X293|A0A1Z2X293_9VIRU Spliced replication-associated protein OS=Bemisia-associated genomovirus NfO OX=1986479 GN=Rep PE=3 SV=2
MM1 pKa = 6.64TTLFFPVKK9 pKa = 9.57STARR13 pKa = 11.84PLKK16 pKa = 8.73MYY18 pKa = 10.28RR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84RR24 pKa = 11.84SAGPRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84TTRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84YY37 pKa = 8.32PVKK40 pKa = 10.28KK41 pKa = 9.4RR42 pKa = 11.84FSRR45 pKa = 11.84RR46 pKa = 11.84TTPTKK51 pKa = 10.13RR52 pKa = 11.84IMRR55 pKa = 11.84RR56 pKa = 11.84TSKK59 pKa = 10.48RR60 pKa = 11.84RR61 pKa = 11.84ILTITSKK68 pKa = 10.71KK69 pKa = 10.27KK70 pKa = 9.73RR71 pKa = 11.84DD72 pKa = 3.48TMLVLTNVQDD82 pKa = 3.49NRR84 pKa = 11.84AITTSYY90 pKa = 10.54NSNGAVMNATAWNGAPWRR108 pKa = 11.84MLWCATARR116 pKa = 11.84NNQNGTGAVGLPDD129 pKa = 3.52NQATRR134 pKa = 11.84TAQACYY140 pKa = 9.71MRR142 pKa = 11.84GLKK145 pKa = 9.55EE146 pKa = 3.79QIEE149 pKa = 5.09FIMNSSLPWQWRR161 pKa = 11.84RR162 pKa = 11.84IVFRR166 pKa = 11.84TKK168 pKa = 10.46GIQAILSEE176 pKa = 4.38TSTSYY181 pKa = 10.87FSNLTSGGYY190 pKa = 10.35ARR192 pKa = 11.84TVNEE196 pKa = 4.57ANTATATNLEE206 pKa = 4.11EE207 pKa = 4.21VLFKK211 pKa = 11.0GKK213 pKa = 9.97QGSDD217 pKa = 2.76WNDD220 pKa = 2.41IFIASTDD227 pKa = 3.41NSRR230 pKa = 11.84VTTMYY235 pKa = 11.2DD236 pKa = 2.86RR237 pKa = 11.84TRR239 pKa = 11.84NLNAGAEE246 pKa = 4.3GGRR249 pKa = 11.84IQKK252 pKa = 9.97VSVWHH257 pKa = 6.27PMNKK261 pKa = 8.23TLVYY265 pKa = 10.05XDD267 pKa = 5.46DD268 pKa = 3.78EE269 pKa = 4.83TGGTEE274 pKa = 3.84ATALYY279 pKa = 9.64SNPSIGLGDD288 pKa = 3.37VWVYY292 pKa = 11.68DD293 pKa = 4.15LFRR296 pKa = 11.84PAYY299 pKa = 8.71GGSGSLVFKK308 pKa = 10.28PQATLYY314 pKa = 8.68WHH316 pKa = 6.98EE317 pKa = 4.28KK318 pKa = 9.3
MM1 pKa = 6.64TTLFFPVKK9 pKa = 9.57STARR13 pKa = 11.84PLKK16 pKa = 8.73MYY18 pKa = 10.28RR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84RR24 pKa = 11.84SAGPRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84TTRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84YY37 pKa = 8.32PVKK40 pKa = 10.28KK41 pKa = 9.4RR42 pKa = 11.84FSRR45 pKa = 11.84RR46 pKa = 11.84TTPTKK51 pKa = 10.13RR52 pKa = 11.84IMRR55 pKa = 11.84RR56 pKa = 11.84TSKK59 pKa = 10.48RR60 pKa = 11.84RR61 pKa = 11.84ILTITSKK68 pKa = 10.71KK69 pKa = 10.27KK70 pKa = 9.73RR71 pKa = 11.84DD72 pKa = 3.48TMLVLTNVQDD82 pKa = 3.49NRR84 pKa = 11.84AITTSYY90 pKa = 10.54NSNGAVMNATAWNGAPWRR108 pKa = 11.84MLWCATARR116 pKa = 11.84NNQNGTGAVGLPDD129 pKa = 3.52NQATRR134 pKa = 11.84TAQACYY140 pKa = 9.71MRR142 pKa = 11.84GLKK145 pKa = 9.55EE146 pKa = 3.79QIEE149 pKa = 5.09FIMNSSLPWQWRR161 pKa = 11.84RR162 pKa = 11.84IVFRR166 pKa = 11.84TKK168 pKa = 10.46GIQAILSEE176 pKa = 4.38TSTSYY181 pKa = 10.87FSNLTSGGYY190 pKa = 10.35ARR192 pKa = 11.84TVNEE196 pKa = 4.57ANTATATNLEE206 pKa = 4.11EE207 pKa = 4.21VLFKK211 pKa = 11.0GKK213 pKa = 9.97QGSDD217 pKa = 2.76WNDD220 pKa = 2.41IFIASTDD227 pKa = 3.41NSRR230 pKa = 11.84VTTMYY235 pKa = 11.2DD236 pKa = 2.86RR237 pKa = 11.84TRR239 pKa = 11.84NLNAGAEE246 pKa = 4.3GGRR249 pKa = 11.84IQKK252 pKa = 9.97VSVWHH257 pKa = 6.27PMNKK261 pKa = 8.23TLVYY265 pKa = 10.05XDD267 pKa = 5.46DD268 pKa = 3.78EE269 pKa = 4.83TGGTEE274 pKa = 3.84ATALYY279 pKa = 9.64SNPSIGLGDD288 pKa = 3.37VWVYY292 pKa = 11.68DD293 pKa = 4.15LFRR296 pKa = 11.84PAYY299 pKa = 8.71GGSGSLVFKK308 pKa = 10.28PQATLYY314 pKa = 8.68WHH316 pKa = 6.98EE317 pKa = 4.28KK318 pKa = 9.3
Molecular weight: 36.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
637 |
318 |
319 |
318.5 |
36.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.064 ± 0.575 | 1.256 ± 0.452 |
5.965 ± 1.808 | 4.553 ± 1.015 |
4.71 ± 1.129 | 8.32 ± 1.011 |
1.727 ± 0.792 | 4.082 ± 0.005 |
4.71 ± 0.459 | 7.535 ± 0.899 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.512 ± 0.456 | 4.396 ± 1.593 |
4.396 ± 0.449 | 2.512 ± 0.456 |
8.948 ± 1.258 | 6.279 ± 0.688 |
8.477 ± 2.504 | 5.651 ± 0.447 |
2.826 ± 0.003 | 3.925 ± 0.109 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
