
Lepus americanus faeces associated genomovirus SHP11
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
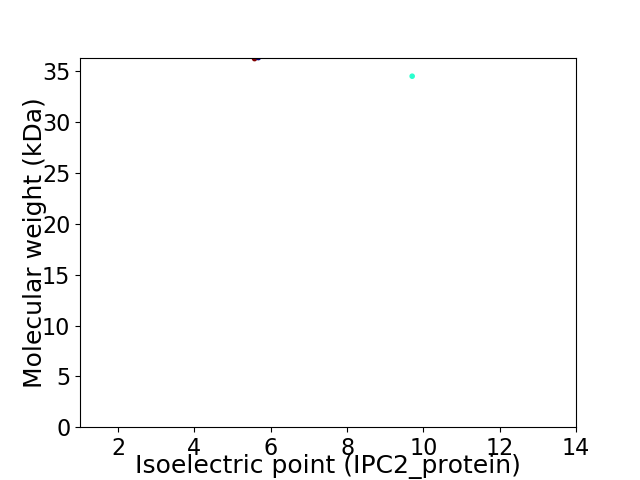
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CJA9|A0A2Z5CJA9_9VIRU Replication-associated protein OS=Lepus americanus faeces associated genomovirus SHP11 OX=2219117 PE=4 SV=1
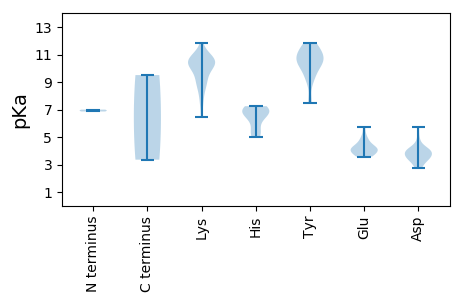
MM1 pKa = 7.0VRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 11.09LEE8 pKa = 3.71EE9 pKa = 3.67VSYY12 pKa = 11.61VLLTYY17 pKa = 7.57PTTPDD22 pKa = 3.06GFDD25 pKa = 3.24PQGIIDD31 pKa = 4.06AVVAAGAVYY40 pKa = 10.25RR41 pKa = 11.84LGRR44 pKa = 11.84EE45 pKa = 3.68LHH47 pKa = 6.88ADD49 pKa = 4.13GKK51 pKa = 8.99PHH53 pKa = 5.26YY54 pKa = 10.02HH55 pKa = 7.07CFVQWADD62 pKa = 3.92PYY64 pKa = 11.14SDD66 pKa = 4.16PDD68 pKa = 3.39AGGTFTVGGRR78 pKa = 11.84RR79 pKa = 11.84PNIKK83 pKa = 10.01KK84 pKa = 10.52FSANPGRR91 pKa = 11.84RR92 pKa = 11.84WDD94 pKa = 3.78YY95 pKa = 10.72VGKK98 pKa = 9.62HH99 pKa = 5.37AGKK102 pKa = 10.38KK103 pKa = 8.76EE104 pKa = 3.58GHH106 pKa = 6.6FIIGDD111 pKa = 3.42QCEE114 pKa = 4.28RR115 pKa = 11.84PSEE118 pKa = 4.14GDD120 pKa = 2.73VDD122 pKa = 3.89TRR124 pKa = 11.84SQSDD128 pKa = 2.78IWSEE132 pKa = 3.99IIGCTTQEE140 pKa = 4.15EE141 pKa = 5.17FWSKK145 pKa = 10.51LAALAPKK152 pKa = 10.22QLGCNFGSLKK162 pKa = 10.8LYY164 pKa = 10.76VDD166 pKa = 4.13WKK168 pKa = 10.36YY169 pKa = 11.42KK170 pKa = 9.0PAQEE174 pKa = 4.7IYY176 pKa = 7.47EE177 pKa = 4.45TPQGTFDD184 pKa = 3.6VPGVLQEE191 pKa = 4.18PKK193 pKa = 10.71GLVLFGPTRR202 pKa = 11.84IGKK205 pKa = 6.42TVWARR210 pKa = 11.84SLADD214 pKa = 3.15HH215 pKa = 7.22AYY217 pKa = 10.45FGGLFNLDD225 pKa = 3.99DD226 pKa = 4.71FSADD230 pKa = 3.03GAQYY234 pKa = 11.09AIFDD238 pKa = 5.15DD239 pKa = 3.83ISGGFSFFPSYY250 pKa = 9.2KK251 pKa = 9.26QWMGGQYY258 pKa = 10.39QFTVTDD264 pKa = 3.91KK265 pKa = 10.76YY266 pKa = 10.28KK267 pKa = 10.97HH268 pKa = 5.95KK269 pKa = 10.37VTLRR273 pKa = 11.84WGRR276 pKa = 11.84PTIWLCNTDD285 pKa = 3.49PRR287 pKa = 11.84EE288 pKa = 3.99DD289 pKa = 3.6HH290 pKa = 6.54YY291 pKa = 11.83KK292 pKa = 10.08PGAMPDD298 pKa = 3.87FAWMEE303 pKa = 4.29EE304 pKa = 3.69NCVFYY309 pKa = 10.62EE310 pKa = 4.02CRR312 pKa = 11.84DD313 pKa = 4.1TIFHH317 pKa = 7.24ASTT320 pKa = 3.35
MM1 pKa = 7.0VRR3 pKa = 11.84RR4 pKa = 11.84FKK6 pKa = 11.09LEE8 pKa = 3.71EE9 pKa = 3.67VSYY12 pKa = 11.61VLLTYY17 pKa = 7.57PTTPDD22 pKa = 3.06GFDD25 pKa = 3.24PQGIIDD31 pKa = 4.06AVVAAGAVYY40 pKa = 10.25RR41 pKa = 11.84LGRR44 pKa = 11.84EE45 pKa = 3.68LHH47 pKa = 6.88ADD49 pKa = 4.13GKK51 pKa = 8.99PHH53 pKa = 5.26YY54 pKa = 10.02HH55 pKa = 7.07CFVQWADD62 pKa = 3.92PYY64 pKa = 11.14SDD66 pKa = 4.16PDD68 pKa = 3.39AGGTFTVGGRR78 pKa = 11.84RR79 pKa = 11.84PNIKK83 pKa = 10.01KK84 pKa = 10.52FSANPGRR91 pKa = 11.84RR92 pKa = 11.84WDD94 pKa = 3.78YY95 pKa = 10.72VGKK98 pKa = 9.62HH99 pKa = 5.37AGKK102 pKa = 10.38KK103 pKa = 8.76EE104 pKa = 3.58GHH106 pKa = 6.6FIIGDD111 pKa = 3.42QCEE114 pKa = 4.28RR115 pKa = 11.84PSEE118 pKa = 4.14GDD120 pKa = 2.73VDD122 pKa = 3.89TRR124 pKa = 11.84SQSDD128 pKa = 2.78IWSEE132 pKa = 3.99IIGCTTQEE140 pKa = 4.15EE141 pKa = 5.17FWSKK145 pKa = 10.51LAALAPKK152 pKa = 10.22QLGCNFGSLKK162 pKa = 10.8LYY164 pKa = 10.76VDD166 pKa = 4.13WKK168 pKa = 10.36YY169 pKa = 11.42KK170 pKa = 9.0PAQEE174 pKa = 4.7IYY176 pKa = 7.47EE177 pKa = 4.45TPQGTFDD184 pKa = 3.6VPGVLQEE191 pKa = 4.18PKK193 pKa = 10.71GLVLFGPTRR202 pKa = 11.84IGKK205 pKa = 6.42TVWARR210 pKa = 11.84SLADD214 pKa = 3.15HH215 pKa = 7.22AYY217 pKa = 10.45FGGLFNLDD225 pKa = 3.99DD226 pKa = 4.71FSADD230 pKa = 3.03GAQYY234 pKa = 11.09AIFDD238 pKa = 5.15DD239 pKa = 3.83ISGGFSFFPSYY250 pKa = 9.2KK251 pKa = 9.26QWMGGQYY258 pKa = 10.39QFTVTDD264 pKa = 3.91KK265 pKa = 10.76YY266 pKa = 10.28KK267 pKa = 10.97HH268 pKa = 5.95KK269 pKa = 10.37VTLRR273 pKa = 11.84WGRR276 pKa = 11.84PTIWLCNTDD285 pKa = 3.49PRR287 pKa = 11.84EE288 pKa = 3.99DD289 pKa = 3.6HH290 pKa = 6.54YY291 pKa = 11.83KK292 pKa = 10.08PGAMPDD298 pKa = 3.87FAWMEE303 pKa = 4.29EE304 pKa = 3.69NCVFYY309 pKa = 10.62EE310 pKa = 4.02CRR312 pKa = 11.84DD313 pKa = 4.1TIFHH317 pKa = 7.24ASTT320 pKa = 3.35
Molecular weight: 36.25 kDa
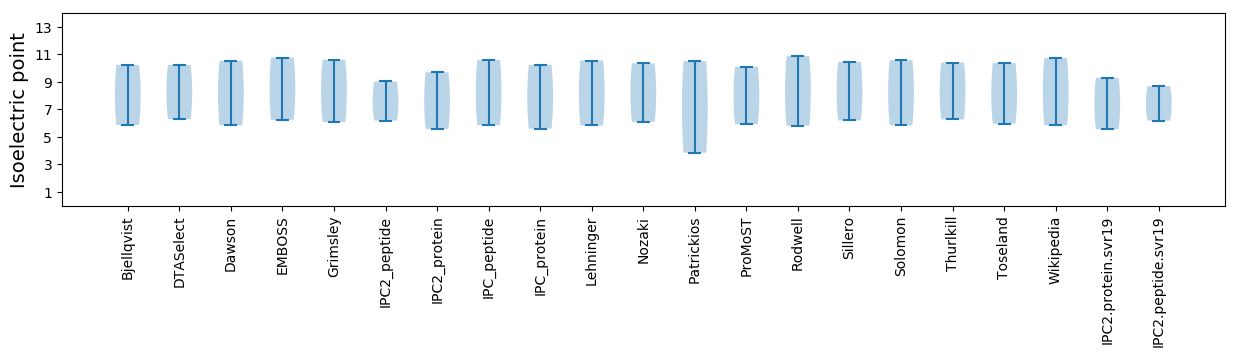
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJA9|A0A2Z5CJA9_9VIRU Replication-associated protein OS=Lepus americanus faeces associated genomovirus SHP11 OX=2219117 PE=4 SV=1
MM1 pKa = 6.86AQRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 8.39FTSYY10 pKa = 9.1RR11 pKa = 11.84AKK13 pKa = 10.33RR14 pKa = 11.84RR15 pKa = 11.84TRR17 pKa = 11.84RR18 pKa = 11.84SARR21 pKa = 11.84PAKK24 pKa = 9.93YY25 pKa = 9.96GGRR28 pKa = 11.84KK29 pKa = 9.1RR30 pKa = 11.84RR31 pKa = 11.84WTKK34 pKa = 7.47TRR36 pKa = 11.84KK37 pKa = 7.67TSRR40 pKa = 11.84PMTKK44 pKa = 10.02KK45 pKa = 10.47RR46 pKa = 11.84ILNTTSRR53 pKa = 11.84KK54 pKa = 9.22KK55 pKa = 10.55RR56 pKa = 11.84NGMLTWSNTNLNGTVRR72 pKa = 11.84PIASGNAYY80 pKa = 9.56VAATNVGVFVYY91 pKa = 10.58CPTALHH97 pKa = 6.86LDD99 pKa = 3.81GDD101 pKa = 4.32SLLRR105 pKa = 11.84NPAARR110 pKa = 11.84TSSTIFAKK118 pKa = 10.6GLSEE122 pKa = 4.2RR123 pKa = 11.84MRR125 pKa = 11.84IQTSSGLPWFHH136 pKa = 6.89RR137 pKa = 11.84RR138 pKa = 11.84VCFTTKK144 pKa = 10.27GPNAFNVINSTDD156 pKa = 3.46TPTVPWAPYY165 pKa = 10.13LDD167 pKa = 3.52TSNGIEE173 pKa = 3.69RR174 pKa = 11.84PMINLDD180 pKa = 3.28INAMPGTLGSQYY192 pKa = 11.63GLLFKK197 pKa = 10.71GRR199 pKa = 11.84QGVDD203 pKa = 2.71WNDD206 pKa = 3.71TIIAPLDD213 pKa = 3.57TARR216 pKa = 11.84VTVKK220 pKa = 10.55YY221 pKa = 10.73DD222 pKa = 3.46KK223 pKa = 10.63TWTLQSGNANGVIRR237 pKa = 11.84EE238 pKa = 4.08RR239 pKa = 11.84KK240 pKa = 8.44LWHH243 pKa = 6.47PMNHH247 pKa = 4.99NVVYY251 pKa = 10.82DD252 pKa = 3.82EE253 pKa = 5.76DD254 pKa = 4.24EE255 pKa = 4.88DD256 pKa = 5.73GEE258 pKa = 4.69VQGTSYY264 pKa = 11.49YY265 pKa = 11.06SVDD268 pKa = 3.46SKK270 pKa = 11.82AGMGDD275 pKa = 3.89YY276 pKa = 10.83YY277 pKa = 11.6VVDD280 pKa = 4.56FFQAGVGGTSTDD292 pKa = 4.19LISVNASSTMYY303 pKa = 9.27WHH305 pKa = 7.08EE306 pKa = 4.08KK307 pKa = 9.5
MM1 pKa = 6.86AQRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 8.39FTSYY10 pKa = 9.1RR11 pKa = 11.84AKK13 pKa = 10.33RR14 pKa = 11.84RR15 pKa = 11.84TRR17 pKa = 11.84RR18 pKa = 11.84SARR21 pKa = 11.84PAKK24 pKa = 9.93YY25 pKa = 9.96GGRR28 pKa = 11.84KK29 pKa = 9.1RR30 pKa = 11.84RR31 pKa = 11.84WTKK34 pKa = 7.47TRR36 pKa = 11.84KK37 pKa = 7.67TSRR40 pKa = 11.84PMTKK44 pKa = 10.02KK45 pKa = 10.47RR46 pKa = 11.84ILNTTSRR53 pKa = 11.84KK54 pKa = 9.22KK55 pKa = 10.55RR56 pKa = 11.84NGMLTWSNTNLNGTVRR72 pKa = 11.84PIASGNAYY80 pKa = 9.56VAATNVGVFVYY91 pKa = 10.58CPTALHH97 pKa = 6.86LDD99 pKa = 3.81GDD101 pKa = 4.32SLLRR105 pKa = 11.84NPAARR110 pKa = 11.84TSSTIFAKK118 pKa = 10.6GLSEE122 pKa = 4.2RR123 pKa = 11.84MRR125 pKa = 11.84IQTSSGLPWFHH136 pKa = 6.89RR137 pKa = 11.84RR138 pKa = 11.84VCFTTKK144 pKa = 10.27GPNAFNVINSTDD156 pKa = 3.46TPTVPWAPYY165 pKa = 10.13LDD167 pKa = 3.52TSNGIEE173 pKa = 3.69RR174 pKa = 11.84PMINLDD180 pKa = 3.28INAMPGTLGSQYY192 pKa = 11.63GLLFKK197 pKa = 10.71GRR199 pKa = 11.84QGVDD203 pKa = 2.71WNDD206 pKa = 3.71TIIAPLDD213 pKa = 3.57TARR216 pKa = 11.84VTVKK220 pKa = 10.55YY221 pKa = 10.73DD222 pKa = 3.46KK223 pKa = 10.63TWTLQSGNANGVIRR237 pKa = 11.84EE238 pKa = 4.08RR239 pKa = 11.84KK240 pKa = 8.44LWHH243 pKa = 6.47PMNHH247 pKa = 4.99NVVYY251 pKa = 10.82DD252 pKa = 3.82EE253 pKa = 5.76DD254 pKa = 4.24EE255 pKa = 4.88DD256 pKa = 5.73GEE258 pKa = 4.69VQGTSYY264 pKa = 11.49YY265 pKa = 11.06SVDD268 pKa = 3.46SKK270 pKa = 11.82AGMGDD275 pKa = 3.89YY276 pKa = 10.83YY277 pKa = 11.6VVDD280 pKa = 4.56FFQAGVGGTSTDD292 pKa = 4.19LISVNASSTMYY303 pKa = 9.27WHH305 pKa = 7.08EE306 pKa = 4.08KK307 pKa = 9.5
Molecular weight: 34.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
627 |
307 |
320 |
313.5 |
35.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.858 ± 0.013 | 1.435 ± 0.577 |
6.539 ± 0.977 | 3.668 ± 1.021 |
4.785 ± 1.363 | 9.091 ± 0.697 |
2.233 ± 0.445 | 4.147 ± 0.175 |
5.901 ± 0.028 | 5.742 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.073 ± 0.631 | 4.147 ± 1.742 |
5.423 ± 0.634 | 3.19 ± 0.669 |
7.018 ± 1.547 | 6.061 ± 1.053 |
8.134 ± 1.684 | 6.061 ± 0.334 |
2.871 ± 0.195 | 4.625 ± 0.287 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
