
Orchid fleck dichorhavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Dichorhavirus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
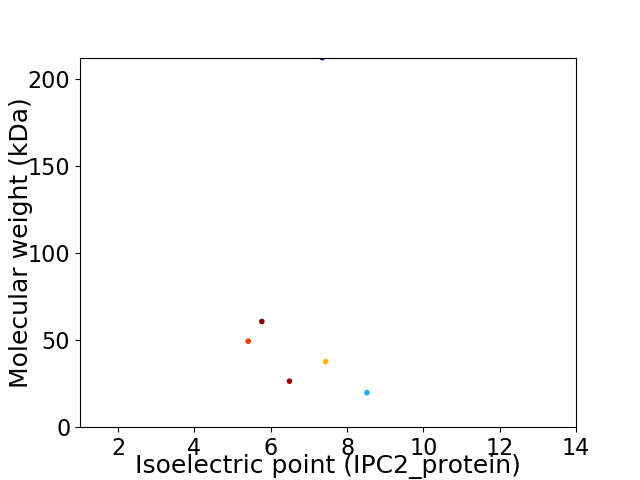
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
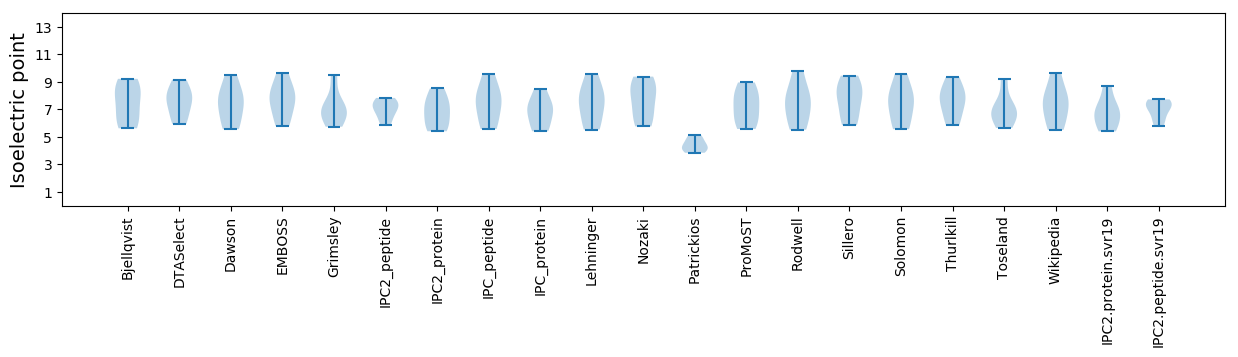
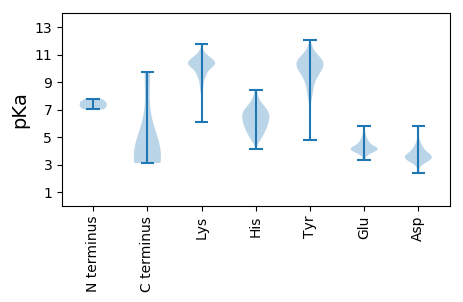
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1MWA2|Q1MWA2_9RHAB Nucleocapsid protein OS=Orchid fleck dichorhavirus OX=152177 GN=N PE=4 SV=1
MM1 pKa = 7.79ANPSEE6 pKa = 4.36IDD8 pKa = 3.35YY9 pKa = 7.46MTPLSAYY16 pKa = 8.51EE17 pKa = 4.13GVPAEE22 pKa = 4.14YY23 pKa = 10.45QEE25 pKa = 4.69ATSSPTPKK33 pKa = 10.37EE34 pKa = 4.06YY35 pKa = 10.43TRR37 pKa = 11.84DD38 pKa = 3.34AAKK41 pKa = 10.53AIPICILPAPPGNEE55 pKa = 3.77VEE57 pKa = 4.22VAEE60 pKa = 4.65AFRR63 pKa = 11.84EE64 pKa = 4.44ATQGTEE70 pKa = 4.01TVLSKK75 pKa = 11.05LQLAQIMSLGFMIQMSGDD93 pKa = 3.79PEE95 pKa = 4.89ALMQGVAEE103 pKa = 4.63GSFLEE108 pKa = 4.7TMTMPDD114 pKa = 2.47IKK116 pKa = 9.92TRR118 pKa = 11.84QDD120 pKa = 3.09SRR122 pKa = 11.84ITAAIAAMALDD133 pKa = 4.13PTEE136 pKa = 4.8EE137 pKa = 4.71GPDD140 pKa = 3.7DD141 pKa = 5.16LEE143 pKa = 5.82DD144 pKa = 4.36RR145 pKa = 11.84SQQADD150 pKa = 3.53AEE152 pKa = 4.59SAMSKK157 pKa = 10.06ARR159 pKa = 11.84AMVYY163 pKa = 9.67ICLSLMRR170 pKa = 11.84LAVKK174 pKa = 9.51PAEE177 pKa = 4.1SFMKK181 pKa = 10.34GVHH184 pKa = 5.75QIKK187 pKa = 9.37QAYY190 pKa = 7.8SVLVGEE196 pKa = 4.56HH197 pKa = 6.54SEE199 pKa = 4.08FLFNYY204 pKa = 9.5SYY206 pKa = 11.55SEE208 pKa = 4.14GMCRR212 pKa = 11.84NIADD216 pKa = 4.46MFNQCDD222 pKa = 3.85DD223 pKa = 4.2LKK225 pKa = 11.07ATLCHH230 pKa = 6.27HH231 pKa = 6.74CAIADD236 pKa = 3.98EE237 pKa = 4.42THH239 pKa = 5.1HH240 pKa = 6.13TNRR243 pKa = 11.84KK244 pKa = 7.81RR245 pKa = 11.84HH246 pKa = 4.7GLLRR250 pKa = 11.84FLILQHH256 pKa = 6.55VDD258 pKa = 3.28LTGMIPYY265 pKa = 10.6GMYY268 pKa = 9.6IDD270 pKa = 3.44MRR272 pKa = 11.84RR273 pKa = 11.84YY274 pKa = 7.85FTLLTPGQLLTWLHH288 pKa = 6.97DD289 pKa = 4.03NQVSRR294 pKa = 11.84PLSVIADD301 pKa = 3.12INTRR305 pKa = 11.84YY306 pKa = 10.11DD307 pKa = 3.19VSNGSDD313 pKa = 3.36RR314 pKa = 11.84FWRR317 pKa = 11.84YY318 pKa = 9.84SRR320 pKa = 11.84GLDD323 pKa = 3.06PGFFIALQQSKK334 pKa = 10.26CVTLIARR341 pKa = 11.84MAHH344 pKa = 6.0ILVKK348 pKa = 10.7GGAVAVNEE356 pKa = 4.04YY357 pKa = 10.9SDD359 pKa = 3.61PRR361 pKa = 11.84KK362 pKa = 10.32AKK364 pKa = 10.46SLEE367 pKa = 4.11NKK369 pKa = 9.62PGLAAEE375 pKa = 4.36ADD377 pKa = 3.53KK378 pKa = 10.78FATEE382 pKa = 4.34FVEE385 pKa = 5.32AYY387 pKa = 9.89NGLSGSSANAGPVSRR402 pKa = 11.84KK403 pKa = 9.75LYY405 pKa = 8.68NQGRR409 pKa = 11.84GIPTRR414 pKa = 11.84RR415 pKa = 11.84GLFTPPSARR424 pKa = 11.84PAPVVNVHH432 pKa = 5.68VPAASSSLTGALDD445 pKa = 4.39AMNSDD450 pKa = 3.92
MM1 pKa = 7.79ANPSEE6 pKa = 4.36IDD8 pKa = 3.35YY9 pKa = 7.46MTPLSAYY16 pKa = 8.51EE17 pKa = 4.13GVPAEE22 pKa = 4.14YY23 pKa = 10.45QEE25 pKa = 4.69ATSSPTPKK33 pKa = 10.37EE34 pKa = 4.06YY35 pKa = 10.43TRR37 pKa = 11.84DD38 pKa = 3.34AAKK41 pKa = 10.53AIPICILPAPPGNEE55 pKa = 3.77VEE57 pKa = 4.22VAEE60 pKa = 4.65AFRR63 pKa = 11.84EE64 pKa = 4.44ATQGTEE70 pKa = 4.01TVLSKK75 pKa = 11.05LQLAQIMSLGFMIQMSGDD93 pKa = 3.79PEE95 pKa = 4.89ALMQGVAEE103 pKa = 4.63GSFLEE108 pKa = 4.7TMTMPDD114 pKa = 2.47IKK116 pKa = 9.92TRR118 pKa = 11.84QDD120 pKa = 3.09SRR122 pKa = 11.84ITAAIAAMALDD133 pKa = 4.13PTEE136 pKa = 4.8EE137 pKa = 4.71GPDD140 pKa = 3.7DD141 pKa = 5.16LEE143 pKa = 5.82DD144 pKa = 4.36RR145 pKa = 11.84SQQADD150 pKa = 3.53AEE152 pKa = 4.59SAMSKK157 pKa = 10.06ARR159 pKa = 11.84AMVYY163 pKa = 9.67ICLSLMRR170 pKa = 11.84LAVKK174 pKa = 9.51PAEE177 pKa = 4.1SFMKK181 pKa = 10.34GVHH184 pKa = 5.75QIKK187 pKa = 9.37QAYY190 pKa = 7.8SVLVGEE196 pKa = 4.56HH197 pKa = 6.54SEE199 pKa = 4.08FLFNYY204 pKa = 9.5SYY206 pKa = 11.55SEE208 pKa = 4.14GMCRR212 pKa = 11.84NIADD216 pKa = 4.46MFNQCDD222 pKa = 3.85DD223 pKa = 4.2LKK225 pKa = 11.07ATLCHH230 pKa = 6.27HH231 pKa = 6.74CAIADD236 pKa = 3.98EE237 pKa = 4.42THH239 pKa = 5.1HH240 pKa = 6.13TNRR243 pKa = 11.84KK244 pKa = 7.81RR245 pKa = 11.84HH246 pKa = 4.7GLLRR250 pKa = 11.84FLILQHH256 pKa = 6.55VDD258 pKa = 3.28LTGMIPYY265 pKa = 10.6GMYY268 pKa = 9.6IDD270 pKa = 3.44MRR272 pKa = 11.84RR273 pKa = 11.84YY274 pKa = 7.85FTLLTPGQLLTWLHH288 pKa = 6.97DD289 pKa = 4.03NQVSRR294 pKa = 11.84PLSVIADD301 pKa = 3.12INTRR305 pKa = 11.84YY306 pKa = 10.11DD307 pKa = 3.19VSNGSDD313 pKa = 3.36RR314 pKa = 11.84FWRR317 pKa = 11.84YY318 pKa = 9.84SRR320 pKa = 11.84GLDD323 pKa = 3.06PGFFIALQQSKK334 pKa = 10.26CVTLIARR341 pKa = 11.84MAHH344 pKa = 6.0ILVKK348 pKa = 10.7GGAVAVNEE356 pKa = 4.04YY357 pKa = 10.9SDD359 pKa = 3.61PRR361 pKa = 11.84KK362 pKa = 10.32AKK364 pKa = 10.46SLEE367 pKa = 4.11NKK369 pKa = 9.62PGLAAEE375 pKa = 4.36ADD377 pKa = 3.53KK378 pKa = 10.78FATEE382 pKa = 4.34FVEE385 pKa = 5.32AYY387 pKa = 9.89NGLSGSSANAGPVSRR402 pKa = 11.84KK403 pKa = 9.75LYY405 pKa = 8.68NQGRR409 pKa = 11.84GIPTRR414 pKa = 11.84RR415 pKa = 11.84GLFTPPSARR424 pKa = 11.84PAPVVNVHH432 pKa = 5.68VPAASSSLTGALDD445 pKa = 4.39AMNSDD450 pKa = 3.92
Molecular weight: 49.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1MWA0|Q1MWA0_9RHAB ORF3 protein OS=Orchid fleck dichorhavirus OX=152177 PE=4 SV=1
MM1 pKa = 7.56SKK3 pKa = 10.18QINMCTVMSFRR14 pKa = 11.84SLITVIGPRR23 pKa = 11.84SVDD26 pKa = 3.07LGAGFKK32 pKa = 10.28EE33 pKa = 4.01AVLRR37 pKa = 11.84AITSLKK43 pKa = 10.6VKK45 pKa = 10.77AKK47 pKa = 9.94DD48 pKa = 3.5PKK50 pKa = 10.41GAEE53 pKa = 3.87IDD55 pKa = 4.4GPWQEE60 pKa = 4.66LLVEE64 pKa = 4.58IATMTKK70 pKa = 10.21SAFTGPTIKK79 pKa = 10.23KK80 pKa = 10.02DD81 pKa = 3.55LEE83 pKa = 3.95QGNVYY88 pKa = 9.87RR89 pKa = 11.84YY90 pKa = 10.27AMTVGGMLSTSHH102 pKa = 5.66NTNGRR107 pKa = 11.84LMTVRR112 pKa = 11.84TGPLYY117 pKa = 10.29DD118 pKa = 3.45TDD120 pKa = 4.25HH121 pKa = 6.27YY122 pKa = 10.69HH123 pKa = 6.69VSGEE127 pKa = 3.9VDD129 pKa = 3.03IDD131 pKa = 3.66KK132 pKa = 10.47MDD134 pKa = 3.72GFKK137 pKa = 10.38ATITLSVATVKK148 pKa = 10.53RR149 pKa = 11.84EE150 pKa = 3.89RR151 pKa = 11.84QQCEE155 pKa = 3.53AGIGEE160 pKa = 4.34YY161 pKa = 10.08HH162 pKa = 6.5VLPIKK167 pKa = 10.56GKK169 pKa = 9.4NPPRR173 pKa = 11.84SASNTAGPSGG183 pKa = 3.38
MM1 pKa = 7.56SKK3 pKa = 10.18QINMCTVMSFRR14 pKa = 11.84SLITVIGPRR23 pKa = 11.84SVDD26 pKa = 3.07LGAGFKK32 pKa = 10.28EE33 pKa = 4.01AVLRR37 pKa = 11.84AITSLKK43 pKa = 10.6VKK45 pKa = 10.77AKK47 pKa = 9.94DD48 pKa = 3.5PKK50 pKa = 10.41GAEE53 pKa = 3.87IDD55 pKa = 4.4GPWQEE60 pKa = 4.66LLVEE64 pKa = 4.58IATMTKK70 pKa = 10.21SAFTGPTIKK79 pKa = 10.23KK80 pKa = 10.02DD81 pKa = 3.55LEE83 pKa = 3.95QGNVYY88 pKa = 9.87RR89 pKa = 11.84YY90 pKa = 10.27AMTVGGMLSTSHH102 pKa = 5.66NTNGRR107 pKa = 11.84LMTVRR112 pKa = 11.84TGPLYY117 pKa = 10.29DD118 pKa = 3.45TDD120 pKa = 4.25HH121 pKa = 6.27YY122 pKa = 10.69HH123 pKa = 6.69VSGEE127 pKa = 3.9VDD129 pKa = 3.03IDD131 pKa = 3.66KK132 pKa = 10.47MDD134 pKa = 3.72GFKK137 pKa = 10.38ATITLSVATVKK148 pKa = 10.53RR149 pKa = 11.84EE150 pKa = 3.89RR151 pKa = 11.84QQCEE155 pKa = 3.53AGIGEE160 pKa = 4.34YY161 pKa = 10.08HH162 pKa = 6.5VLPIKK167 pKa = 10.56GKK169 pKa = 9.4NPPRR173 pKa = 11.84SASNTAGPSGG183 pKa = 3.38
Molecular weight: 19.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3624 |
183 |
1877 |
604.0 |
67.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.015 ± 1.206 | 2.07 ± 0.265 |
5.464 ± 0.412 | 5.602 ± 0.276 |
2.98 ± 0.284 | 6.485 ± 0.284 |
2.925 ± 0.41 | 6.319 ± 0.49 |
5.85 ± 0.449 | 8.609 ± 0.486 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.305 ± 0.36 | 4.222 ± 0.166 |
4.691 ± 0.59 | 2.732 ± 0.324 |
5.16 ± 0.538 | 8.223 ± 0.278 |
6.705 ± 0.769 | 6.843 ± 0.524 |
1.738 ± 0.329 | 3.063 ± 0.278 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
