
Odonata-associated circular virus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Dragsmacovirus; Dragonfly associated dragsmacovirus 1
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
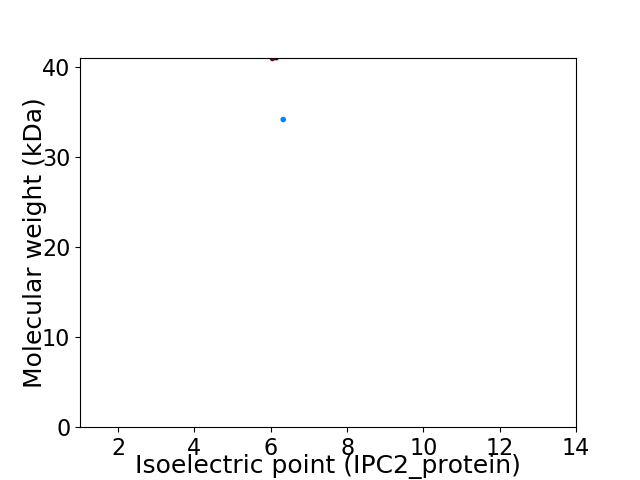
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI30|A0A0B4UI30_9VIRU Putative capsid protein OS=Odonata-associated circular virus 5 OX=1592125 PE=4 SV=1
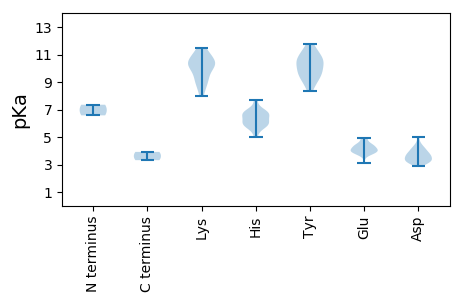
MM1 pKa = 6.58TTQFAQASYY10 pKa = 11.25QEE12 pKa = 4.91IIDD15 pKa = 4.59LNTVTDD21 pKa = 3.88KK22 pKa = 11.47VSVLGIHH29 pKa = 6.06TPVTDD34 pKa = 3.52TPYY37 pKa = 10.8TFLKK41 pKa = 10.56PFFDD45 pKa = 4.44AFQKK49 pKa = 10.26YY50 pKa = 9.78HH51 pKa = 6.82YY52 pKa = 9.01DD53 pKa = 3.61GCSLTMVPAARR64 pKa = 11.84LPADD68 pKa = 4.18PSQVSYY74 pKa = 11.24EE75 pKa = 4.24SGEE78 pKa = 3.99QPIDD82 pKa = 3.51PRR84 pKa = 11.84DD85 pKa = 3.77LLNPILWHH93 pKa = 5.94GAHH96 pKa = 6.59GEE98 pKa = 4.17SLGAVLNQFYY108 pKa = 10.25DD109 pKa = 3.64ASGSTSDD116 pKa = 3.23IGRR119 pKa = 11.84EE120 pKa = 3.86FFDD123 pKa = 3.23SGEE126 pKa = 4.05VSRR129 pKa = 11.84ASLTQVGNDD138 pKa = 3.32ALYY141 pKa = 10.24EE142 pKa = 4.1SLYY145 pKa = 10.78YY146 pKa = 10.17RR147 pKa = 11.84ALTDD151 pKa = 3.47NSWRR155 pKa = 11.84KK156 pKa = 8.79AHH158 pKa = 6.44PQTGLRR164 pKa = 11.84VKK166 pKa = 10.28GLHH169 pKa = 6.07PLVYY173 pKa = 10.45SVGTDD178 pKa = 3.12AQYY181 pKa = 11.78LNNPTGTTTPQVPRR195 pKa = 11.84SANSPLPPSDD205 pKa = 4.26AGPMTSVGAAGTMGKK220 pKa = 8.35GTSSPYY226 pKa = 10.79GLQSLNPQGLQDD238 pKa = 3.78TSDD241 pKa = 3.96SGDD244 pKa = 3.28LTYY247 pKa = 10.92VRR249 pKa = 11.84SLPTPRR255 pKa = 11.84TSQFFTEE262 pKa = 4.4RR263 pKa = 11.84LHH265 pKa = 6.68GLGWLDD271 pKa = 3.12TRR273 pKa = 11.84VRR275 pKa = 11.84ISSTSGFQAEE285 pKa = 4.45VSGNQTLDD293 pKa = 2.9RR294 pKa = 11.84MAVEE298 pKa = 4.04NLYY301 pKa = 10.59EE302 pKa = 4.01QVEE305 pKa = 4.39DD306 pKa = 3.78SQTINYY312 pKa = 9.09LPRR315 pKa = 11.84LFMGVCLLPPSYY327 pKa = 10.12KK328 pKa = 10.29AKK330 pKa = 9.93QYY332 pKa = 9.55YY333 pKa = 9.81RR334 pKa = 11.84LVINHH339 pKa = 6.76RR340 pKa = 11.84FSFRR344 pKa = 11.84KK345 pKa = 9.56FRR347 pKa = 11.84GLSMDD352 pKa = 3.11GGLRR356 pKa = 11.84GRR358 pKa = 11.84YY359 pKa = 9.04GPARR363 pKa = 11.84APSYY367 pKa = 10.67RR368 pKa = 11.84NLMM371 pKa = 3.92
MM1 pKa = 6.58TTQFAQASYY10 pKa = 11.25QEE12 pKa = 4.91IIDD15 pKa = 4.59LNTVTDD21 pKa = 3.88KK22 pKa = 11.47VSVLGIHH29 pKa = 6.06TPVTDD34 pKa = 3.52TPYY37 pKa = 10.8TFLKK41 pKa = 10.56PFFDD45 pKa = 4.44AFQKK49 pKa = 10.26YY50 pKa = 9.78HH51 pKa = 6.82YY52 pKa = 9.01DD53 pKa = 3.61GCSLTMVPAARR64 pKa = 11.84LPADD68 pKa = 4.18PSQVSYY74 pKa = 11.24EE75 pKa = 4.24SGEE78 pKa = 3.99QPIDD82 pKa = 3.51PRR84 pKa = 11.84DD85 pKa = 3.77LLNPILWHH93 pKa = 5.94GAHH96 pKa = 6.59GEE98 pKa = 4.17SLGAVLNQFYY108 pKa = 10.25DD109 pKa = 3.64ASGSTSDD116 pKa = 3.23IGRR119 pKa = 11.84EE120 pKa = 3.86FFDD123 pKa = 3.23SGEE126 pKa = 4.05VSRR129 pKa = 11.84ASLTQVGNDD138 pKa = 3.32ALYY141 pKa = 10.24EE142 pKa = 4.1SLYY145 pKa = 10.78YY146 pKa = 10.17RR147 pKa = 11.84ALTDD151 pKa = 3.47NSWRR155 pKa = 11.84KK156 pKa = 8.79AHH158 pKa = 6.44PQTGLRR164 pKa = 11.84VKK166 pKa = 10.28GLHH169 pKa = 6.07PLVYY173 pKa = 10.45SVGTDD178 pKa = 3.12AQYY181 pKa = 11.78LNNPTGTTTPQVPRR195 pKa = 11.84SANSPLPPSDD205 pKa = 4.26AGPMTSVGAAGTMGKK220 pKa = 8.35GTSSPYY226 pKa = 10.79GLQSLNPQGLQDD238 pKa = 3.78TSDD241 pKa = 3.96SGDD244 pKa = 3.28LTYY247 pKa = 10.92VRR249 pKa = 11.84SLPTPRR255 pKa = 11.84TSQFFTEE262 pKa = 4.4RR263 pKa = 11.84LHH265 pKa = 6.68GLGWLDD271 pKa = 3.12TRR273 pKa = 11.84VRR275 pKa = 11.84ISSTSGFQAEE285 pKa = 4.45VSGNQTLDD293 pKa = 2.9RR294 pKa = 11.84MAVEE298 pKa = 4.04NLYY301 pKa = 10.59EE302 pKa = 4.01QVEE305 pKa = 4.39DD306 pKa = 3.78SQTINYY312 pKa = 9.09LPRR315 pKa = 11.84LFMGVCLLPPSYY327 pKa = 10.12KK328 pKa = 10.29AKK330 pKa = 9.93QYY332 pKa = 9.55YY333 pKa = 9.81RR334 pKa = 11.84LVINHH339 pKa = 6.76RR340 pKa = 11.84FSFRR344 pKa = 11.84KK345 pKa = 9.56FRR347 pKa = 11.84GLSMDD352 pKa = 3.11GGLRR356 pKa = 11.84GRR358 pKa = 11.84YY359 pKa = 9.04GPARR363 pKa = 11.84APSYY367 pKa = 10.67RR368 pKa = 11.84NLMM371 pKa = 3.92
Molecular weight: 40.93 kDa
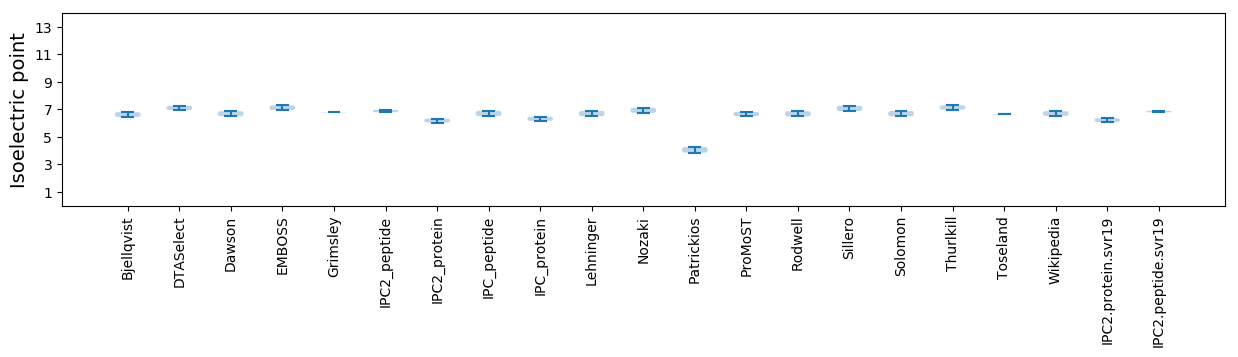
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI30|A0A0B4UI30_9VIRU Putative capsid protein OS=Odonata-associated circular virus 5 OX=1592125 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.57WSIYY6 pKa = 9.67IICGPIRR13 pKa = 11.84IVIRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 10.46RR20 pKa = 11.84SMTEE24 pKa = 3.96PKK26 pKa = 9.25WWDD29 pKa = 3.24CTLSRR34 pKa = 11.84EE35 pKa = 4.24EE36 pKa = 4.31TFGNGNPNPYY46 pKa = 8.35YY47 pKa = 9.39TWHH50 pKa = 6.74APDD53 pKa = 3.31VGKK56 pKa = 10.26QLMEE60 pKa = 4.95HH61 pKa = 5.79GAEE64 pKa = 3.68RR65 pKa = 11.84CVVGEE70 pKa = 3.99EE71 pKa = 4.11VGEE74 pKa = 4.35DD75 pKa = 4.11GYY77 pKa = 10.51QHH79 pKa = 5.77FQIRR83 pKa = 11.84VVFKK87 pKa = 10.79KK88 pKa = 8.84PTSFEE93 pKa = 3.84KK94 pKa = 10.23ATAILPGSHH103 pKa = 6.21WSEE106 pKa = 3.74TSQFGRR112 pKa = 11.84NFDD115 pKa = 3.81YY116 pKa = 11.28VEE118 pKa = 4.46KK119 pKa = 10.5EE120 pKa = 3.85GHH122 pKa = 5.91FWRR125 pKa = 11.84SWEE128 pKa = 4.07TALGRR133 pKa = 11.84FMQLDD138 pKa = 4.03MYY140 pKa = 9.9PWQNEE145 pKa = 3.09IMTRR149 pKa = 11.84LEE151 pKa = 3.88KK152 pKa = 11.04QNDD155 pKa = 3.73RR156 pKa = 11.84KK157 pKa = 11.12VMVIVDD163 pKa = 4.02RR164 pKa = 11.84YY165 pKa = 9.53GNSGKK170 pKa = 8.01TAIAMRR176 pKa = 11.84LTAEE180 pKa = 4.48HH181 pKa = 6.77KK182 pKa = 10.43GAYY185 pKa = 9.7CPEE188 pKa = 4.92LEE190 pKa = 4.71DD191 pKa = 5.04AKK193 pKa = 11.38DD194 pKa = 3.65YY195 pKa = 8.56MAWALAHH202 pKa = 6.89KK203 pKa = 10.34NAGCFCLDD211 pKa = 3.14IPRR214 pKa = 11.84AGDD217 pKa = 3.34VRR219 pKa = 11.84KK220 pKa = 9.03DD221 pKa = 3.24TAIWKK226 pKa = 9.82AVEE229 pKa = 3.91QMKK232 pKa = 10.71NGYY235 pKa = 9.2LWDD238 pKa = 4.31KK239 pKa = 9.19RR240 pKa = 11.84HH241 pKa = 5.79HH242 pKa = 5.03WQEE245 pKa = 3.73AFIMPSKK252 pKa = 10.94GLVLTNDD259 pKa = 3.35EE260 pKa = 4.43PDD262 pKa = 3.49RR263 pKa = 11.84NLLSRR268 pKa = 11.84DD269 pKa = 2.98RR270 pKa = 11.84WDD272 pKa = 4.13IGHH275 pKa = 7.69LDD277 pKa = 3.58YY278 pKa = 11.53GISNWANTIKK288 pKa = 10.11WDD290 pKa = 3.62SS291 pKa = 3.32
MM1 pKa = 7.35KK2 pKa = 9.57WSIYY6 pKa = 9.67IICGPIRR13 pKa = 11.84IVIRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 10.46RR20 pKa = 11.84SMTEE24 pKa = 3.96PKK26 pKa = 9.25WWDD29 pKa = 3.24CTLSRR34 pKa = 11.84EE35 pKa = 4.24EE36 pKa = 4.31TFGNGNPNPYY46 pKa = 8.35YY47 pKa = 9.39TWHH50 pKa = 6.74APDD53 pKa = 3.31VGKK56 pKa = 10.26QLMEE60 pKa = 4.95HH61 pKa = 5.79GAEE64 pKa = 3.68RR65 pKa = 11.84CVVGEE70 pKa = 3.99EE71 pKa = 4.11VGEE74 pKa = 4.35DD75 pKa = 4.11GYY77 pKa = 10.51QHH79 pKa = 5.77FQIRR83 pKa = 11.84VVFKK87 pKa = 10.79KK88 pKa = 8.84PTSFEE93 pKa = 3.84KK94 pKa = 10.23ATAILPGSHH103 pKa = 6.21WSEE106 pKa = 3.74TSQFGRR112 pKa = 11.84NFDD115 pKa = 3.81YY116 pKa = 11.28VEE118 pKa = 4.46KK119 pKa = 10.5EE120 pKa = 3.85GHH122 pKa = 5.91FWRR125 pKa = 11.84SWEE128 pKa = 4.07TALGRR133 pKa = 11.84FMQLDD138 pKa = 4.03MYY140 pKa = 9.9PWQNEE145 pKa = 3.09IMTRR149 pKa = 11.84LEE151 pKa = 3.88KK152 pKa = 11.04QNDD155 pKa = 3.73RR156 pKa = 11.84KK157 pKa = 11.12VMVIVDD163 pKa = 4.02RR164 pKa = 11.84YY165 pKa = 9.53GNSGKK170 pKa = 8.01TAIAMRR176 pKa = 11.84LTAEE180 pKa = 4.48HH181 pKa = 6.77KK182 pKa = 10.43GAYY185 pKa = 9.7CPEE188 pKa = 4.92LEE190 pKa = 4.71DD191 pKa = 5.04AKK193 pKa = 11.38DD194 pKa = 3.65YY195 pKa = 8.56MAWALAHH202 pKa = 6.89KK203 pKa = 10.34NAGCFCLDD211 pKa = 3.14IPRR214 pKa = 11.84AGDD217 pKa = 3.34VRR219 pKa = 11.84KK220 pKa = 9.03DD221 pKa = 3.24TAIWKK226 pKa = 9.82AVEE229 pKa = 3.91QMKK232 pKa = 10.71NGYY235 pKa = 9.2LWDD238 pKa = 4.31KK239 pKa = 9.19RR240 pKa = 11.84HH241 pKa = 5.79HH242 pKa = 5.03WQEE245 pKa = 3.73AFIMPSKK252 pKa = 10.94GLVLTNDD259 pKa = 3.35EE260 pKa = 4.43PDD262 pKa = 3.49RR263 pKa = 11.84NLLSRR268 pKa = 11.84DD269 pKa = 2.98RR270 pKa = 11.84WDD272 pKa = 4.13IGHH275 pKa = 7.69LDD277 pKa = 3.58YY278 pKa = 11.53GISNWANTIKK288 pKa = 10.11WDD290 pKa = 3.62SS291 pKa = 3.32
Molecular weight: 34.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
662 |
291 |
371 |
331.0 |
37.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.344 ± 0.119 | 1.208 ± 0.547 |
6.193 ± 0.215 | 4.985 ± 1.432 |
3.776 ± 0.218 | 8.006 ± 0.507 |
2.719 ± 0.46 | 3.927 ± 1.228 |
4.23 ± 1.475 | 8.006 ± 1.609 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.87 ± 0.584 | 4.079 ± 0.249 |
5.891 ± 1.134 | 4.381 ± 0.826 |
6.495 ± 0.242 | 7.402 ± 1.883 |
6.647 ± 1.178 | 5.287 ± 0.305 |
2.719 ± 1.563 | 4.834 ± 0.456 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
