
Piscine myocarditis virus AL V-708
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
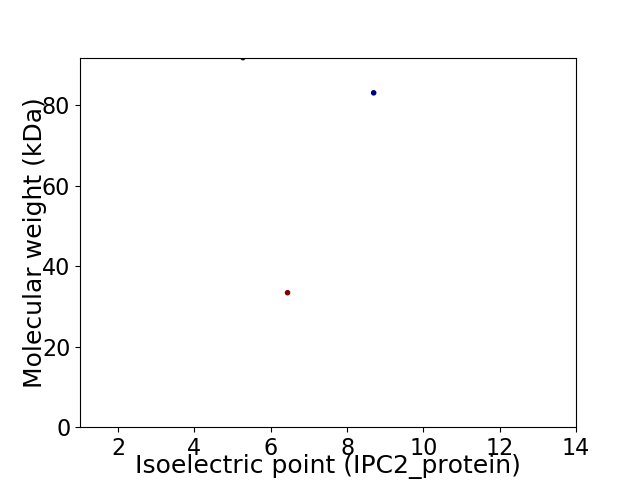
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
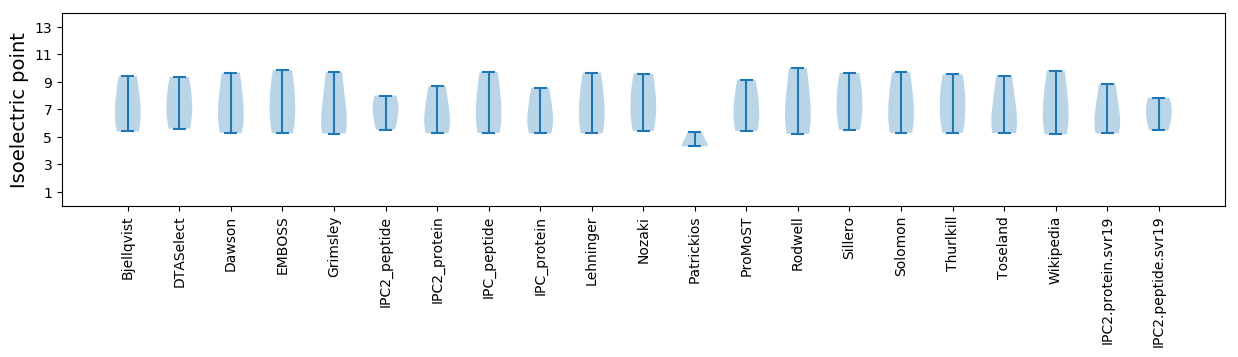
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E3W912|E3W912_9VIRU RNA-directed RNA polymerase OS=Piscine myocarditis virus AL V-708 OX=912320 PE=3 SV=1
MM1 pKa = 7.66EE2 pKa = 6.22PNTSVIATEE11 pKa = 4.01QQQAAMRR18 pKa = 11.84EE19 pKa = 4.41VEE21 pKa = 4.29AEE23 pKa = 3.5AAARR27 pKa = 11.84DD28 pKa = 3.8EE29 pKa = 4.39VVEE32 pKa = 4.8KK33 pKa = 10.67IAFAEE38 pKa = 4.48GAMMVQTRR46 pKa = 11.84RR47 pKa = 11.84LPSGKK52 pKa = 9.96SSVGGFLGEE61 pKa = 4.05LAQNIRR67 pKa = 11.84AMNRR71 pKa = 11.84SLHH74 pKa = 6.23TDD76 pKa = 2.82TNMLTEE82 pKa = 4.11GAMVDD87 pKa = 3.33RR88 pKa = 11.84ARR90 pKa = 11.84AKK92 pKa = 7.48VHH94 pKa = 6.15KK95 pKa = 9.94IIRR98 pKa = 11.84EE99 pKa = 4.08GNLDD103 pKa = 3.4SRR105 pKa = 11.84VFSNTGSNTMLSLWVPAVPGPPAVPEE131 pKa = 4.15HH132 pKa = 6.92WDD134 pKa = 3.43VAPSWFVCRR143 pKa = 11.84PGKK146 pKa = 10.37KK147 pKa = 10.03GGIKK151 pKa = 8.63ITQSASMAALNPLFRR166 pKa = 11.84GADD169 pKa = 3.29VGPIGTAVRR178 pKa = 11.84ADD180 pKa = 3.44VNAFSMNAVLGALRR194 pKa = 11.84AGGFNTEE201 pKa = 3.76HH202 pKa = 6.68SLVSFVEE209 pKa = 3.76PLIRR213 pKa = 11.84ILLMGVQTQDD223 pKa = 3.08RR224 pKa = 11.84GTSPWDD230 pKa = 3.15WVGGMSSRR238 pKa = 11.84IVNPLVFTTSGNFFPGGPNLRR259 pKa = 11.84VWGANDD265 pKa = 3.22TVARR269 pKa = 11.84IVNVEE274 pKa = 3.83DD275 pKa = 3.75YY276 pKa = 9.78MRR278 pKa = 11.84EE279 pKa = 3.75AAGEE283 pKa = 4.22GRR285 pKa = 11.84FDD287 pKa = 4.39AGWGPEE293 pKa = 4.04FWGGTGDD300 pKa = 3.79DD301 pKa = 3.85AVAVVPIRR309 pKa = 11.84AVEE312 pKa = 4.09AGLGEE317 pKa = 4.57VNAGWTLAHH326 pKa = 6.36MEE328 pKa = 4.33YY329 pKa = 9.87PVKK332 pKa = 10.77VRR334 pKa = 11.84LLDD337 pKa = 3.54VDD339 pKa = 4.46DD340 pKa = 4.01RR341 pKa = 11.84TIGPGGSLPLNANRR355 pKa = 11.84EE356 pKa = 4.11YY357 pKa = 9.54TAAGATHH364 pKa = 6.4VPGPYY369 pKa = 10.13ARR371 pKa = 11.84VLYY374 pKa = 10.5VVVDD378 pKa = 4.38QNADD382 pKa = 3.33RR383 pKa = 11.84CVGVRR388 pKa = 11.84VQGQGAVIDD397 pKa = 3.88VDD399 pKa = 3.56PALNYY404 pKa = 10.48VIGGADD410 pKa = 3.85LGMLPLIQWSVGLGAEE426 pKa = 4.59DD427 pKa = 3.66MAQGSIAQTQRR438 pKa = 11.84WVRR441 pKa = 11.84MYY443 pKa = 11.13GNEE446 pKa = 4.68DD447 pKa = 3.17DD448 pKa = 5.49WEE450 pKa = 4.69SAWHH454 pKa = 5.9LVSSAYY460 pKa = 8.9TVYY463 pKa = 10.78SPAFRR468 pKa = 11.84RR469 pKa = 11.84SGVAVEE475 pKa = 4.44GGFWAQPAAGAAPFPLGGLAGWVRR499 pKa = 11.84YY500 pKa = 9.8DD501 pKa = 3.47NQARR505 pKa = 11.84AAQVALCRR513 pKa = 11.84EE514 pKa = 4.13RR515 pKa = 11.84ADD517 pKa = 3.51MAEE520 pKa = 4.54CPWGGYY526 pKa = 7.07RR527 pKa = 11.84EE528 pKa = 4.04RR529 pKa = 11.84GVRR532 pKa = 11.84PGSVANWQYY541 pKa = 11.32VRR543 pKa = 11.84FDD545 pKa = 3.28PTVAVGVAAHH555 pKa = 6.47FWSVVKK561 pKa = 10.73VMVAPVPDD569 pKa = 3.78RR570 pKa = 11.84AAALADD576 pKa = 3.76MAWGKK581 pKa = 11.07GKK583 pKa = 9.66VQAMGEE589 pKa = 4.14DD590 pKa = 4.19VINGQMGQPEE600 pKa = 4.69SMMRR604 pKa = 11.84GVALNEE610 pKa = 4.07NQGLAAATVRR620 pKa = 11.84RR621 pKa = 11.84VVGLEE626 pKa = 3.87NEE628 pKa = 4.59SMQTTHH634 pKa = 6.25WSTTEE639 pKa = 3.66VAMNGYY645 pKa = 9.34YY646 pKa = 10.55GRR648 pKa = 11.84AGATAHH654 pKa = 6.63HH655 pKa = 6.67AAFPLSEE662 pKa = 4.39GGTMRR667 pKa = 11.84KK668 pKa = 9.79RR669 pKa = 11.84IPAIEE674 pKa = 3.84MRR676 pKa = 11.84EE677 pKa = 3.97NGVEE681 pKa = 4.0GDD683 pKa = 4.31LMNDD687 pKa = 3.43DD688 pKa = 5.37LYY690 pKa = 11.61SIGTAAGYY698 pKa = 9.76LAVEE702 pKa = 4.79GMAGAQGGIWDD713 pKa = 3.86VVQYY717 pKa = 9.15QLPGPDD723 pKa = 3.42DD724 pKa = 3.6EE725 pKa = 5.87ARR727 pKa = 11.84GVMNTVGAMGGWTRR741 pKa = 11.84AVTPVDD747 pKa = 3.4NVATMRR753 pKa = 11.84DD754 pKa = 3.33NGVEE758 pKa = 4.01GEE760 pKa = 4.12PCGIVMSLPTSGTAVVDD777 pKa = 4.04RR778 pKa = 11.84LANFGLPPARR788 pKa = 11.84AEE790 pKa = 3.92LRR792 pKa = 11.84EE793 pKa = 4.14VPFGGYY799 pKa = 7.64QRR801 pKa = 11.84SVTNTNHH808 pKa = 5.89RR809 pKa = 11.84VKK811 pKa = 10.85VSVSGGRR818 pKa = 11.84AVVQKK823 pKa = 10.92GNKK826 pKa = 9.86AEE828 pKa = 4.05MNPVFVNRR836 pKa = 11.84TPGQTTLGQPTTDD849 pKa = 3.28TTGMTTADD857 pKa = 3.79FLDD860 pKa = 3.58II861 pKa = 4.88
MM1 pKa = 7.66EE2 pKa = 6.22PNTSVIATEE11 pKa = 4.01QQQAAMRR18 pKa = 11.84EE19 pKa = 4.41VEE21 pKa = 4.29AEE23 pKa = 3.5AAARR27 pKa = 11.84DD28 pKa = 3.8EE29 pKa = 4.39VVEE32 pKa = 4.8KK33 pKa = 10.67IAFAEE38 pKa = 4.48GAMMVQTRR46 pKa = 11.84RR47 pKa = 11.84LPSGKK52 pKa = 9.96SSVGGFLGEE61 pKa = 4.05LAQNIRR67 pKa = 11.84AMNRR71 pKa = 11.84SLHH74 pKa = 6.23TDD76 pKa = 2.82TNMLTEE82 pKa = 4.11GAMVDD87 pKa = 3.33RR88 pKa = 11.84ARR90 pKa = 11.84AKK92 pKa = 7.48VHH94 pKa = 6.15KK95 pKa = 9.94IIRR98 pKa = 11.84EE99 pKa = 4.08GNLDD103 pKa = 3.4SRR105 pKa = 11.84VFSNTGSNTMLSLWVPAVPGPPAVPEE131 pKa = 4.15HH132 pKa = 6.92WDD134 pKa = 3.43VAPSWFVCRR143 pKa = 11.84PGKK146 pKa = 10.37KK147 pKa = 10.03GGIKK151 pKa = 8.63ITQSASMAALNPLFRR166 pKa = 11.84GADD169 pKa = 3.29VGPIGTAVRR178 pKa = 11.84ADD180 pKa = 3.44VNAFSMNAVLGALRR194 pKa = 11.84AGGFNTEE201 pKa = 3.76HH202 pKa = 6.68SLVSFVEE209 pKa = 3.76PLIRR213 pKa = 11.84ILLMGVQTQDD223 pKa = 3.08RR224 pKa = 11.84GTSPWDD230 pKa = 3.15WVGGMSSRR238 pKa = 11.84IVNPLVFTTSGNFFPGGPNLRR259 pKa = 11.84VWGANDD265 pKa = 3.22TVARR269 pKa = 11.84IVNVEE274 pKa = 3.83DD275 pKa = 3.75YY276 pKa = 9.78MRR278 pKa = 11.84EE279 pKa = 3.75AAGEE283 pKa = 4.22GRR285 pKa = 11.84FDD287 pKa = 4.39AGWGPEE293 pKa = 4.04FWGGTGDD300 pKa = 3.79DD301 pKa = 3.85AVAVVPIRR309 pKa = 11.84AVEE312 pKa = 4.09AGLGEE317 pKa = 4.57VNAGWTLAHH326 pKa = 6.36MEE328 pKa = 4.33YY329 pKa = 9.87PVKK332 pKa = 10.77VRR334 pKa = 11.84LLDD337 pKa = 3.54VDD339 pKa = 4.46DD340 pKa = 4.01RR341 pKa = 11.84TIGPGGSLPLNANRR355 pKa = 11.84EE356 pKa = 4.11YY357 pKa = 9.54TAAGATHH364 pKa = 6.4VPGPYY369 pKa = 10.13ARR371 pKa = 11.84VLYY374 pKa = 10.5VVVDD378 pKa = 4.38QNADD382 pKa = 3.33RR383 pKa = 11.84CVGVRR388 pKa = 11.84VQGQGAVIDD397 pKa = 3.88VDD399 pKa = 3.56PALNYY404 pKa = 10.48VIGGADD410 pKa = 3.85LGMLPLIQWSVGLGAEE426 pKa = 4.59DD427 pKa = 3.66MAQGSIAQTQRR438 pKa = 11.84WVRR441 pKa = 11.84MYY443 pKa = 11.13GNEE446 pKa = 4.68DD447 pKa = 3.17DD448 pKa = 5.49WEE450 pKa = 4.69SAWHH454 pKa = 5.9LVSSAYY460 pKa = 8.9TVYY463 pKa = 10.78SPAFRR468 pKa = 11.84RR469 pKa = 11.84SGVAVEE475 pKa = 4.44GGFWAQPAAGAAPFPLGGLAGWVRR499 pKa = 11.84YY500 pKa = 9.8DD501 pKa = 3.47NQARR505 pKa = 11.84AAQVALCRR513 pKa = 11.84EE514 pKa = 4.13RR515 pKa = 11.84ADD517 pKa = 3.51MAEE520 pKa = 4.54CPWGGYY526 pKa = 7.07RR527 pKa = 11.84EE528 pKa = 4.04RR529 pKa = 11.84GVRR532 pKa = 11.84PGSVANWQYY541 pKa = 11.32VRR543 pKa = 11.84FDD545 pKa = 3.28PTVAVGVAAHH555 pKa = 6.47FWSVVKK561 pKa = 10.73VMVAPVPDD569 pKa = 3.78RR570 pKa = 11.84AAALADD576 pKa = 3.76MAWGKK581 pKa = 11.07GKK583 pKa = 9.66VQAMGEE589 pKa = 4.14DD590 pKa = 4.19VINGQMGQPEE600 pKa = 4.69SMMRR604 pKa = 11.84GVALNEE610 pKa = 4.07NQGLAAATVRR620 pKa = 11.84RR621 pKa = 11.84VVGLEE626 pKa = 3.87NEE628 pKa = 4.59SMQTTHH634 pKa = 6.25WSTTEE639 pKa = 3.66VAMNGYY645 pKa = 9.34YY646 pKa = 10.55GRR648 pKa = 11.84AGATAHH654 pKa = 6.63HH655 pKa = 6.67AAFPLSEE662 pKa = 4.39GGTMRR667 pKa = 11.84KK668 pKa = 9.79RR669 pKa = 11.84IPAIEE674 pKa = 3.84MRR676 pKa = 11.84EE677 pKa = 3.97NGVEE681 pKa = 4.0GDD683 pKa = 4.31LMNDD687 pKa = 3.43DD688 pKa = 5.37LYY690 pKa = 11.61SIGTAAGYY698 pKa = 9.76LAVEE702 pKa = 4.79GMAGAQGGIWDD713 pKa = 3.86VVQYY717 pKa = 9.15QLPGPDD723 pKa = 3.42DD724 pKa = 3.6EE725 pKa = 5.87ARR727 pKa = 11.84GVMNTVGAMGGWTRR741 pKa = 11.84AVTPVDD747 pKa = 3.4NVATMRR753 pKa = 11.84DD754 pKa = 3.33NGVEE758 pKa = 4.01GEE760 pKa = 4.12PCGIVMSLPTSGTAVVDD777 pKa = 4.04RR778 pKa = 11.84LANFGLPPARR788 pKa = 11.84AEE790 pKa = 3.92LRR792 pKa = 11.84EE793 pKa = 4.14VPFGGYY799 pKa = 7.64QRR801 pKa = 11.84SVTNTNHH808 pKa = 5.89RR809 pKa = 11.84VKK811 pKa = 10.85VSVSGGRR818 pKa = 11.84AVVQKK823 pKa = 10.92GNKK826 pKa = 9.86AEE828 pKa = 4.05MNPVFVNRR836 pKa = 11.84TPGQTTLGQPTTDD849 pKa = 3.28TTGMTTADD857 pKa = 3.79FLDD860 pKa = 3.58II861 pKa = 4.88
Molecular weight: 91.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3W913|E3W913_9VIRU Structural protein OS=Piscine myocarditis virus AL V-708 OX=912320 PE=4 SV=1
MM1 pKa = 6.82ITKK4 pKa = 10.3ALAQGRR10 pKa = 11.84EE11 pKa = 4.04EE12 pKa = 4.16DD13 pKa = 3.76WIKK16 pKa = 9.9ITTNTTKK23 pKa = 10.35IKK25 pKa = 10.47LKK27 pKa = 9.7SLRR30 pKa = 11.84PLDD33 pKa = 3.81GWEE36 pKa = 3.82HH37 pKa = 6.1SGWGLGLRR45 pKa = 11.84LALEE49 pKa = 4.46LVSEE53 pKa = 4.64PTRR56 pKa = 11.84EE57 pKa = 4.03TMSGVIRR64 pKa = 11.84QIRR67 pKa = 11.84IGKK70 pKa = 9.4LDD72 pKa = 3.39PTKK75 pKa = 10.67VFKK78 pKa = 10.93SISNQAKK85 pKa = 9.57VSGCGLCRR93 pKa = 11.84EE94 pKa = 4.14WKK96 pKa = 10.37EE97 pKa = 3.92YY98 pKa = 10.72VNWEE102 pKa = 4.12LLGGYY107 pKa = 8.03QMITAQQMGEE117 pKa = 4.19EE118 pKa = 4.48VAHH121 pKa = 5.94QVTEE125 pKa = 4.13KK126 pKa = 10.87AKK128 pKa = 10.59SEE130 pKa = 4.04EE131 pKa = 4.2AGSDD135 pKa = 3.5RR136 pKa = 11.84INRR139 pKa = 11.84ALEE142 pKa = 4.52SILGPLRR149 pKa = 11.84GTCTVPCSQRR159 pKa = 11.84EE160 pKa = 4.0FLLCRR165 pKa = 11.84DD166 pKa = 3.28KK167 pKa = 10.57WAKK170 pKa = 8.19NTSGYY175 pKa = 10.08IGLGQNLGRR184 pKa = 11.84QKK186 pKa = 10.52IDD188 pKa = 2.96VALNSDD194 pKa = 3.42IKK196 pKa = 11.0ALEE199 pKa = 3.74RR200 pKa = 11.84MMEE203 pKa = 4.12GEE205 pKa = 4.08FEE207 pKa = 4.0NRR209 pKa = 11.84PFIKK213 pKa = 10.28SEE215 pKa = 4.03PGKK218 pKa = 10.48SRR220 pKa = 11.84PVVNSNISCYY230 pKa = 10.92LNLEE234 pKa = 4.42YY235 pKa = 10.95GWVTIKK241 pKa = 9.75KK242 pKa = 7.39TLKK245 pKa = 10.11KK246 pKa = 10.41YY247 pKa = 9.85LGKK250 pKa = 9.22RR251 pKa = 11.84TTIFDD256 pKa = 3.91DD257 pKa = 4.23ASQKK261 pKa = 10.02VLLWQEE267 pKa = 4.12MIIGADD273 pKa = 3.15NHH275 pKa = 6.87RR276 pKa = 11.84EE277 pKa = 3.84IKK279 pKa = 10.89VPLDD283 pKa = 3.14YY284 pKa = 11.37SRR286 pKa = 11.84FDD288 pKa = 3.31STIGKK293 pKa = 5.64TQIMKK298 pKa = 10.17CVRR301 pKa = 11.84LLLDD305 pKa = 3.32MVRR308 pKa = 11.84GNDD311 pKa = 2.75NWKK314 pKa = 10.27RR315 pKa = 11.84SVTEE319 pKa = 3.55RR320 pKa = 11.84FEE322 pKa = 4.12RR323 pKa = 11.84QTVYY327 pKa = 10.53IDD329 pKa = 3.95GYY331 pKa = 11.59GKK333 pKa = 10.28LRR335 pKa = 11.84WEE337 pKa = 4.17NSVLSGWRR345 pKa = 11.84WTSIITSVINLAILKK360 pKa = 8.58ATGADD365 pKa = 3.29ASGVGIKK372 pKa = 10.26VQGDD376 pKa = 3.55DD377 pKa = 3.94VKK379 pKa = 11.12ISFQTIRR386 pKa = 11.84EE387 pKa = 4.18AEE389 pKa = 4.08RR390 pKa = 11.84CVEE393 pKa = 4.3EE394 pKa = 4.55INALGYY400 pKa = 9.78EE401 pKa = 4.13INPGKK406 pKa = 10.68VFASRR411 pKa = 11.84KK412 pKa = 7.51RR413 pKa = 11.84DD414 pKa = 3.1EE415 pKa = 4.4YY416 pKa = 11.26LRR418 pKa = 11.84MVAEE422 pKa = 4.38EE423 pKa = 4.52GKK425 pKa = 8.95MAGYY429 pKa = 10.09VIRR432 pKa = 11.84CLPKK436 pKa = 9.92IVFTSPTEE444 pKa = 4.08EE445 pKa = 3.78VTTWEE450 pKa = 3.67EE451 pKa = 4.2RR452 pKa = 11.84IRR454 pKa = 11.84GTVSKK459 pKa = 9.97WMRR462 pKa = 11.84VLSRR466 pKa = 11.84GGDD469 pKa = 3.46RR470 pKa = 11.84EE471 pKa = 4.19VAKK474 pKa = 10.98YY475 pKa = 8.6WMKK478 pKa = 10.88RR479 pKa = 11.84DD480 pKa = 3.45LCGLSGEE487 pKa = 4.13NSEE490 pKa = 6.53LIDD493 pKa = 3.2SWLRR497 pKa = 11.84TPASVGGGGCAWLLGEE513 pKa = 4.51GNLWTGLVLRR523 pKa = 11.84EE524 pKa = 3.94QEE526 pKa = 3.85EE527 pKa = 4.64AEE529 pKa = 4.13VVEE532 pKa = 4.39RR533 pKa = 11.84SNHH536 pKa = 5.48KK537 pKa = 10.09MMSDD541 pKa = 3.57YY542 pKa = 11.01GGSVPARR549 pKa = 11.84RR550 pKa = 11.84WIKK553 pKa = 10.77SVTKK557 pKa = 9.9GAKK560 pKa = 8.87PAFGFKK566 pKa = 10.01RR567 pKa = 11.84IEE569 pKa = 3.99RR570 pKa = 11.84VKK572 pKa = 10.88PLGNWLKK579 pKa = 10.65DD580 pKa = 3.44LRR582 pKa = 11.84PVAEE586 pKa = 4.48SIKK589 pKa = 10.66SISDD593 pKa = 3.57LVEE596 pKa = 3.92TSQVPKK602 pKa = 10.6FRR604 pKa = 11.84FKK606 pKa = 10.63EE607 pKa = 4.04IYY609 pKa = 9.65DD610 pKa = 3.3WTPFMVHH617 pKa = 5.93KK618 pKa = 10.53RR619 pKa = 11.84NALIEE624 pKa = 4.2EE625 pKa = 4.41KK626 pKa = 10.52NWDD629 pKa = 3.99KK630 pKa = 11.42LCSLYY635 pKa = 11.0HH636 pKa = 6.07NEE638 pKa = 4.5GEE640 pKa = 4.04VRR642 pKa = 11.84LYY644 pKa = 11.03KK645 pKa = 10.45RR646 pKa = 11.84ILPRR650 pKa = 11.84WMWLSMIKK658 pKa = 10.37NEE660 pKa = 4.16LTWVSMPPNIANSEE674 pKa = 4.26VASLAKK680 pKa = 10.24EE681 pKa = 3.67RR682 pKa = 11.84VEE684 pKa = 3.94YY685 pKa = 10.55KK686 pKa = 10.66VLSKK690 pKa = 10.37VLRR693 pKa = 11.84YY694 pKa = 9.44HH695 pKa = 5.68RR696 pKa = 11.84TVSRR700 pKa = 11.84EE701 pKa = 3.46TLTRR705 pKa = 11.84FSIGGEE711 pKa = 4.06YY712 pKa = 10.43VLSAKK717 pKa = 10.71LNDD720 pKa = 3.73RR721 pKa = 11.84QRR723 pKa = 11.84ILQQ726 pKa = 3.73
MM1 pKa = 6.82ITKK4 pKa = 10.3ALAQGRR10 pKa = 11.84EE11 pKa = 4.04EE12 pKa = 4.16DD13 pKa = 3.76WIKK16 pKa = 9.9ITTNTTKK23 pKa = 10.35IKK25 pKa = 10.47LKK27 pKa = 9.7SLRR30 pKa = 11.84PLDD33 pKa = 3.81GWEE36 pKa = 3.82HH37 pKa = 6.1SGWGLGLRR45 pKa = 11.84LALEE49 pKa = 4.46LVSEE53 pKa = 4.64PTRR56 pKa = 11.84EE57 pKa = 4.03TMSGVIRR64 pKa = 11.84QIRR67 pKa = 11.84IGKK70 pKa = 9.4LDD72 pKa = 3.39PTKK75 pKa = 10.67VFKK78 pKa = 10.93SISNQAKK85 pKa = 9.57VSGCGLCRR93 pKa = 11.84EE94 pKa = 4.14WKK96 pKa = 10.37EE97 pKa = 3.92YY98 pKa = 10.72VNWEE102 pKa = 4.12LLGGYY107 pKa = 8.03QMITAQQMGEE117 pKa = 4.19EE118 pKa = 4.48VAHH121 pKa = 5.94QVTEE125 pKa = 4.13KK126 pKa = 10.87AKK128 pKa = 10.59SEE130 pKa = 4.04EE131 pKa = 4.2AGSDD135 pKa = 3.5RR136 pKa = 11.84INRR139 pKa = 11.84ALEE142 pKa = 4.52SILGPLRR149 pKa = 11.84GTCTVPCSQRR159 pKa = 11.84EE160 pKa = 4.0FLLCRR165 pKa = 11.84DD166 pKa = 3.28KK167 pKa = 10.57WAKK170 pKa = 8.19NTSGYY175 pKa = 10.08IGLGQNLGRR184 pKa = 11.84QKK186 pKa = 10.52IDD188 pKa = 2.96VALNSDD194 pKa = 3.42IKK196 pKa = 11.0ALEE199 pKa = 3.74RR200 pKa = 11.84MMEE203 pKa = 4.12GEE205 pKa = 4.08FEE207 pKa = 4.0NRR209 pKa = 11.84PFIKK213 pKa = 10.28SEE215 pKa = 4.03PGKK218 pKa = 10.48SRR220 pKa = 11.84PVVNSNISCYY230 pKa = 10.92LNLEE234 pKa = 4.42YY235 pKa = 10.95GWVTIKK241 pKa = 9.75KK242 pKa = 7.39TLKK245 pKa = 10.11KK246 pKa = 10.41YY247 pKa = 9.85LGKK250 pKa = 9.22RR251 pKa = 11.84TTIFDD256 pKa = 3.91DD257 pKa = 4.23ASQKK261 pKa = 10.02VLLWQEE267 pKa = 4.12MIIGADD273 pKa = 3.15NHH275 pKa = 6.87RR276 pKa = 11.84EE277 pKa = 3.84IKK279 pKa = 10.89VPLDD283 pKa = 3.14YY284 pKa = 11.37SRR286 pKa = 11.84FDD288 pKa = 3.31STIGKK293 pKa = 5.64TQIMKK298 pKa = 10.17CVRR301 pKa = 11.84LLLDD305 pKa = 3.32MVRR308 pKa = 11.84GNDD311 pKa = 2.75NWKK314 pKa = 10.27RR315 pKa = 11.84SVTEE319 pKa = 3.55RR320 pKa = 11.84FEE322 pKa = 4.12RR323 pKa = 11.84QTVYY327 pKa = 10.53IDD329 pKa = 3.95GYY331 pKa = 11.59GKK333 pKa = 10.28LRR335 pKa = 11.84WEE337 pKa = 4.17NSVLSGWRR345 pKa = 11.84WTSIITSVINLAILKK360 pKa = 8.58ATGADD365 pKa = 3.29ASGVGIKK372 pKa = 10.26VQGDD376 pKa = 3.55DD377 pKa = 3.94VKK379 pKa = 11.12ISFQTIRR386 pKa = 11.84EE387 pKa = 4.18AEE389 pKa = 4.08RR390 pKa = 11.84CVEE393 pKa = 4.3EE394 pKa = 4.55INALGYY400 pKa = 9.78EE401 pKa = 4.13INPGKK406 pKa = 10.68VFASRR411 pKa = 11.84KK412 pKa = 7.51RR413 pKa = 11.84DD414 pKa = 3.1EE415 pKa = 4.4YY416 pKa = 11.26LRR418 pKa = 11.84MVAEE422 pKa = 4.38EE423 pKa = 4.52GKK425 pKa = 8.95MAGYY429 pKa = 10.09VIRR432 pKa = 11.84CLPKK436 pKa = 9.92IVFTSPTEE444 pKa = 4.08EE445 pKa = 3.78VTTWEE450 pKa = 3.67EE451 pKa = 4.2RR452 pKa = 11.84IRR454 pKa = 11.84GTVSKK459 pKa = 9.97WMRR462 pKa = 11.84VLSRR466 pKa = 11.84GGDD469 pKa = 3.46RR470 pKa = 11.84EE471 pKa = 4.19VAKK474 pKa = 10.98YY475 pKa = 8.6WMKK478 pKa = 10.88RR479 pKa = 11.84DD480 pKa = 3.45LCGLSGEE487 pKa = 4.13NSEE490 pKa = 6.53LIDD493 pKa = 3.2SWLRR497 pKa = 11.84TPASVGGGGCAWLLGEE513 pKa = 4.51GNLWTGLVLRR523 pKa = 11.84EE524 pKa = 3.94QEE526 pKa = 3.85EE527 pKa = 4.64AEE529 pKa = 4.13VVEE532 pKa = 4.39RR533 pKa = 11.84SNHH536 pKa = 5.48KK537 pKa = 10.09MMSDD541 pKa = 3.57YY542 pKa = 11.01GGSVPARR549 pKa = 11.84RR550 pKa = 11.84WIKK553 pKa = 10.77SVTKK557 pKa = 9.9GAKK560 pKa = 8.87PAFGFKK566 pKa = 10.01RR567 pKa = 11.84IEE569 pKa = 3.99RR570 pKa = 11.84VKK572 pKa = 10.88PLGNWLKK579 pKa = 10.65DD580 pKa = 3.44LRR582 pKa = 11.84PVAEE586 pKa = 4.48SIKK589 pKa = 10.66SISDD593 pKa = 3.57LVEE596 pKa = 3.92TSQVPKK602 pKa = 10.6FRR604 pKa = 11.84FKK606 pKa = 10.63EE607 pKa = 4.04IYY609 pKa = 9.65DD610 pKa = 3.3WTPFMVHH617 pKa = 5.93KK618 pKa = 10.53RR619 pKa = 11.84NALIEE624 pKa = 4.2EE625 pKa = 4.41KK626 pKa = 10.52NWDD629 pKa = 3.99KK630 pKa = 11.42LCSLYY635 pKa = 11.0HH636 pKa = 6.07NEE638 pKa = 4.5GEE640 pKa = 4.04VRR642 pKa = 11.84LYY644 pKa = 11.03KK645 pKa = 10.45RR646 pKa = 11.84ILPRR650 pKa = 11.84WMWLSMIKK658 pKa = 10.37NEE660 pKa = 4.16LTWVSMPPNIANSEE674 pKa = 4.26VASLAKK680 pKa = 10.24EE681 pKa = 3.67RR682 pKa = 11.84VEE684 pKa = 3.94YY685 pKa = 10.55KK686 pKa = 10.66VLSKK690 pKa = 10.37VLRR693 pKa = 11.84YY694 pKa = 9.44HH695 pKa = 5.68RR696 pKa = 11.84TVSRR700 pKa = 11.84EE701 pKa = 3.46TLTRR705 pKa = 11.84FSIGGEE711 pKa = 4.06YY712 pKa = 10.43VLSAKK717 pKa = 10.71LNDD720 pKa = 3.73RR721 pKa = 11.84QRR723 pKa = 11.84ILQQ726 pKa = 3.73
Molecular weight: 83.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1889 |
302 |
861 |
629.7 |
69.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.994 ± 2.506 | 1.535 ± 0.682 |
4.394 ± 0.369 | 7.253 ± 1.138 |
2.594 ± 0.243 | 10.323 ± 1.362 |
1.323 ± 0.225 | 4.87 ± 1.206 |
5.188 ± 2.078 | 7.517 ± 1.137 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.282 ± 0.537 | 4.553 ± 0.355 |
4.288 ± 0.813 | 3.123 ± 0.286 |
6.988 ± 0.607 | 5.664 ± 0.849 |
5.188 ± 0.479 | 8.947 ± 1.293 |
2.647 ± 0.523 | 2.329 ± 0.234 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
