
Acinetobacter sp. CAG:196
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Acinetobacter; environmental samples
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
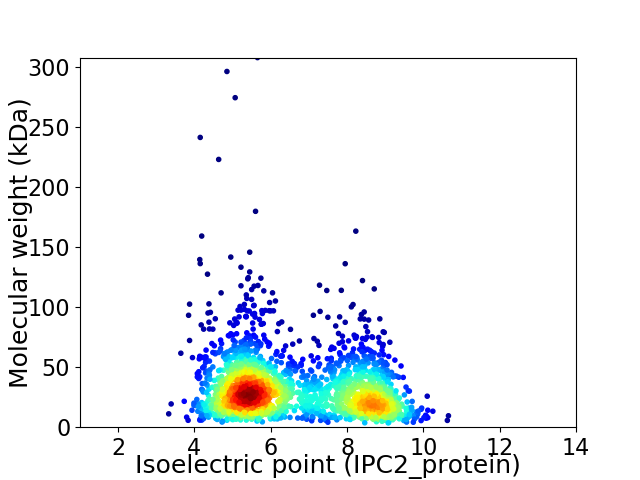
Virtual 2D-PAGE plot for 2085 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
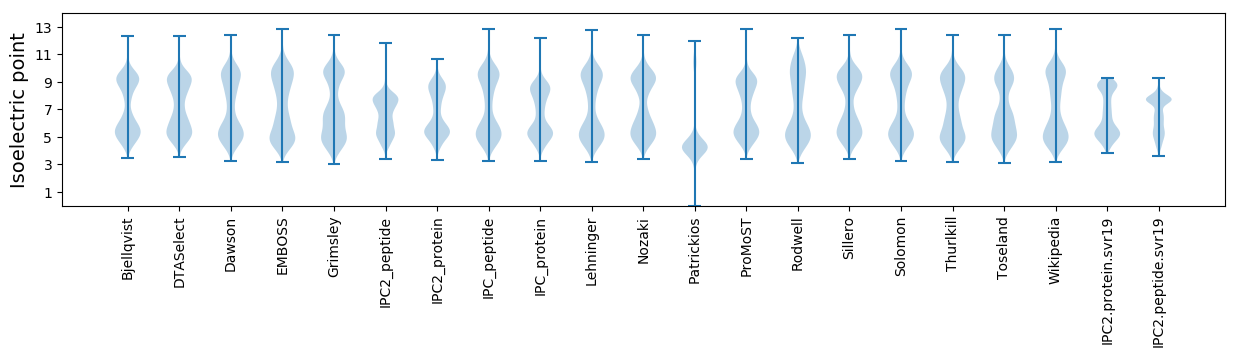
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5SHP1|R5SHP1_9GAMM Uncharacterized protein OS=Acinetobacter sp. CAG:196 OX=1262690 GN=BN527_00523 PE=4 SV=1
MM1 pKa = 7.39GNDD4 pKa = 3.27IEE6 pKa = 4.36SLQNILKK13 pKa = 9.99NDD15 pKa = 3.59PSNFQARR22 pKa = 11.84RR23 pKa = 11.84EE24 pKa = 3.94LSILLVNNGFNEE36 pKa = 4.11EE37 pKa = 4.16AEE39 pKa = 4.5GNLKK43 pKa = 10.46YY44 pKa = 10.56LLKK47 pKa = 10.72YY48 pKa = 10.24FPQDD52 pKa = 2.92AEE54 pKa = 3.81IYY56 pKa = 8.27YY57 pKa = 10.59NLGIVYY63 pKa = 9.19EE64 pKa = 4.1KK65 pKa = 10.73LKK67 pKa = 10.96KK68 pKa = 8.91FTEE71 pKa = 4.34AKK73 pKa = 9.56EE74 pKa = 4.42CYY76 pKa = 9.02QKK78 pKa = 11.04AVEE81 pKa = 4.71ISPQEE86 pKa = 3.73DD87 pKa = 3.92FYY89 pKa = 12.01YY90 pKa = 11.12NLGDD94 pKa = 3.55VLVDD98 pKa = 3.9LKK100 pKa = 10.99EE101 pKa = 3.75WDD103 pKa = 3.58EE104 pKa = 4.76AISAFRR110 pKa = 11.84KK111 pKa = 10.0VLEE114 pKa = 4.41TDD116 pKa = 3.71PEE118 pKa = 4.68DD119 pKa = 3.91GNCYY123 pKa = 10.14FNLGVCYY130 pKa = 10.28FNKK133 pKa = 10.23DD134 pKa = 3.68EE135 pKa = 5.64KK136 pKa = 11.35NLALDD141 pKa = 4.04NFQKK145 pKa = 10.87AAAINPKK152 pKa = 10.41DD153 pKa = 3.58VFAYY157 pKa = 9.46FYY159 pKa = 11.05LGFIYY164 pKa = 10.74QNDD167 pKa = 4.02GLTNFAIDD175 pKa = 3.48SYY177 pKa = 11.46KK178 pKa = 10.47KK179 pKa = 10.4VLEE182 pKa = 4.04ISPDD186 pKa = 3.52YY187 pKa = 10.87SWAYY191 pKa = 10.5FNLGSIAYY199 pKa = 8.93KK200 pKa = 10.26SGNIEE205 pKa = 3.7EE206 pKa = 4.23AKK208 pKa = 10.44EE209 pKa = 3.89YY210 pKa = 10.78LLKK213 pKa = 9.98TLEE216 pKa = 4.26YY217 pKa = 10.16NSSDD221 pKa = 2.93IEE223 pKa = 4.22AYY225 pKa = 9.75KK226 pKa = 10.89LLTKK230 pKa = 10.13ICLKK234 pKa = 10.04TGEE237 pKa = 4.4TEE239 pKa = 4.28EE240 pKa = 4.16ILEE243 pKa = 4.15ILNTRR248 pKa = 11.84LDD250 pKa = 3.69KK251 pKa = 11.13EE252 pKa = 4.72DD253 pKa = 4.48NGDD256 pKa = 4.01LYY258 pKa = 11.63YY259 pKa = 11.12CLARR263 pKa = 11.84VYY265 pKa = 10.55KK266 pKa = 10.55YY267 pKa = 10.49IGDD270 pKa = 3.83SEE272 pKa = 4.68QYY274 pKa = 10.48CKK276 pKa = 10.71NLEE279 pKa = 4.03QALDD283 pKa = 3.62NPYY286 pKa = 9.25TLTFSKK292 pKa = 10.52KK293 pKa = 9.01IVKK296 pKa = 9.85QEE298 pKa = 3.69LEE300 pKa = 4.42YY301 pKa = 10.57IKK303 pKa = 11.04EE304 pKa = 4.0MLGRR308 pKa = 11.84NAVIEE313 pKa = 4.14PEE315 pKa = 4.0AGIEE319 pKa = 4.24EE320 pKa = 4.47YY321 pKa = 10.7SEE323 pKa = 4.19EE324 pKa = 4.13EE325 pKa = 4.15PEE327 pKa = 3.85EE328 pKa = 4.29DD329 pKa = 3.98EE330 pKa = 4.9NYY332 pKa = 10.5EE333 pKa = 4.99DD334 pKa = 3.77EE335 pKa = 5.18PEE337 pKa = 4.3YY338 pKa = 11.13DD339 pKa = 4.52DD340 pKa = 5.4EE341 pKa = 6.76ADD343 pKa = 3.67EE344 pKa = 4.45YY345 pKa = 11.44EE346 pKa = 4.18EE347 pKa = 6.29DD348 pKa = 4.53EE349 pKa = 6.15DD350 pKa = 5.06YY351 pKa = 11.66DD352 pKa = 5.09DD353 pKa = 6.49DD354 pKa = 5.76GEE356 pKa = 4.17EE357 pKa = 4.37DD358 pKa = 3.53SS359 pKa = 5.06
MM1 pKa = 7.39GNDD4 pKa = 3.27IEE6 pKa = 4.36SLQNILKK13 pKa = 9.99NDD15 pKa = 3.59PSNFQARR22 pKa = 11.84RR23 pKa = 11.84EE24 pKa = 3.94LSILLVNNGFNEE36 pKa = 4.11EE37 pKa = 4.16AEE39 pKa = 4.5GNLKK43 pKa = 10.46YY44 pKa = 10.56LLKK47 pKa = 10.72YY48 pKa = 10.24FPQDD52 pKa = 2.92AEE54 pKa = 3.81IYY56 pKa = 8.27YY57 pKa = 10.59NLGIVYY63 pKa = 9.19EE64 pKa = 4.1KK65 pKa = 10.73LKK67 pKa = 10.96KK68 pKa = 8.91FTEE71 pKa = 4.34AKK73 pKa = 9.56EE74 pKa = 4.42CYY76 pKa = 9.02QKK78 pKa = 11.04AVEE81 pKa = 4.71ISPQEE86 pKa = 3.73DD87 pKa = 3.92FYY89 pKa = 12.01YY90 pKa = 11.12NLGDD94 pKa = 3.55VLVDD98 pKa = 3.9LKK100 pKa = 10.99EE101 pKa = 3.75WDD103 pKa = 3.58EE104 pKa = 4.76AISAFRR110 pKa = 11.84KK111 pKa = 10.0VLEE114 pKa = 4.41TDD116 pKa = 3.71PEE118 pKa = 4.68DD119 pKa = 3.91GNCYY123 pKa = 10.14FNLGVCYY130 pKa = 10.28FNKK133 pKa = 10.23DD134 pKa = 3.68EE135 pKa = 5.64KK136 pKa = 11.35NLALDD141 pKa = 4.04NFQKK145 pKa = 10.87AAAINPKK152 pKa = 10.41DD153 pKa = 3.58VFAYY157 pKa = 9.46FYY159 pKa = 11.05LGFIYY164 pKa = 10.74QNDD167 pKa = 4.02GLTNFAIDD175 pKa = 3.48SYY177 pKa = 11.46KK178 pKa = 10.47KK179 pKa = 10.4VLEE182 pKa = 4.04ISPDD186 pKa = 3.52YY187 pKa = 10.87SWAYY191 pKa = 10.5FNLGSIAYY199 pKa = 8.93KK200 pKa = 10.26SGNIEE205 pKa = 3.7EE206 pKa = 4.23AKK208 pKa = 10.44EE209 pKa = 3.89YY210 pKa = 10.78LLKK213 pKa = 9.98TLEE216 pKa = 4.26YY217 pKa = 10.16NSSDD221 pKa = 2.93IEE223 pKa = 4.22AYY225 pKa = 9.75KK226 pKa = 10.89LLTKK230 pKa = 10.13ICLKK234 pKa = 10.04TGEE237 pKa = 4.4TEE239 pKa = 4.28EE240 pKa = 4.16ILEE243 pKa = 4.15ILNTRR248 pKa = 11.84LDD250 pKa = 3.69KK251 pKa = 11.13EE252 pKa = 4.72DD253 pKa = 4.48NGDD256 pKa = 4.01LYY258 pKa = 11.63YY259 pKa = 11.12CLARR263 pKa = 11.84VYY265 pKa = 10.55KK266 pKa = 10.55YY267 pKa = 10.49IGDD270 pKa = 3.83SEE272 pKa = 4.68QYY274 pKa = 10.48CKK276 pKa = 10.71NLEE279 pKa = 4.03QALDD283 pKa = 3.62NPYY286 pKa = 9.25TLTFSKK292 pKa = 10.52KK293 pKa = 9.01IVKK296 pKa = 9.85QEE298 pKa = 3.69LEE300 pKa = 4.42YY301 pKa = 10.57IKK303 pKa = 11.04EE304 pKa = 4.0MLGRR308 pKa = 11.84NAVIEE313 pKa = 4.14PEE315 pKa = 4.0AGIEE319 pKa = 4.24EE320 pKa = 4.47YY321 pKa = 10.7SEE323 pKa = 4.19EE324 pKa = 4.13EE325 pKa = 4.15PEE327 pKa = 3.85EE328 pKa = 4.29DD329 pKa = 3.98EE330 pKa = 4.9NYY332 pKa = 10.5EE333 pKa = 4.99DD334 pKa = 3.77EE335 pKa = 5.18PEE337 pKa = 4.3YY338 pKa = 11.13DD339 pKa = 4.52DD340 pKa = 5.4EE341 pKa = 6.76ADD343 pKa = 3.67EE344 pKa = 4.45YY345 pKa = 11.44EE346 pKa = 4.18EE347 pKa = 6.29DD348 pKa = 4.53EE349 pKa = 6.15DD350 pKa = 5.06YY351 pKa = 11.66DD352 pKa = 5.09DD353 pKa = 6.49DD354 pKa = 5.76GEE356 pKa = 4.17EE357 pKa = 4.37DD358 pKa = 3.53SS359 pKa = 5.06
Molecular weight: 41.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5SSV7|R5SSV7_9GAMM Pyruvate formate-lyase-activating enzyme OS=Acinetobacter sp. CAG:196 OX=1262690 GN=BN527_02002 PE=3 SV=1
MM1 pKa = 7.37ARR3 pKa = 11.84LVGVDD8 pKa = 3.47LPRR11 pKa = 11.84NKK13 pKa = 10.1RR14 pKa = 11.84LEE16 pKa = 3.77IALTYY21 pKa = 10.09IYY23 pKa = 10.47GIGPARR29 pKa = 11.84SKK31 pKa = 10.88KK32 pKa = 9.0ILEE35 pKa = 4.24VTGISPDD42 pKa = 3.68LRR44 pKa = 11.84TDD46 pKa = 3.84DD47 pKa = 5.1LTDD50 pKa = 4.72DD51 pKa = 5.67DD52 pKa = 5.33IKK54 pKa = 10.99QLRR57 pKa = 11.84NALSEE62 pKa = 4.15YY63 pKa = 10.54NIEE66 pKa = 3.93GDD68 pKa = 3.53LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.58VTMNIKK78 pKa = 10.21RR79 pKa = 11.84LQEE82 pKa = 3.81INSYY86 pKa = 10.63RR87 pKa = 11.84GMRR90 pKa = 11.84HH91 pKa = 6.02KK92 pKa = 10.66RR93 pKa = 11.84GLPCRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 10.84TNARR107 pKa = 11.84TRR109 pKa = 11.84RR110 pKa = 11.84GKK112 pKa = 8.91KK113 pKa = 7.98TGPIAGKK120 pKa = 10.3KK121 pKa = 8.86KK122 pKa = 10.5
MM1 pKa = 7.37ARR3 pKa = 11.84LVGVDD8 pKa = 3.47LPRR11 pKa = 11.84NKK13 pKa = 10.1RR14 pKa = 11.84LEE16 pKa = 3.77IALTYY21 pKa = 10.09IYY23 pKa = 10.47GIGPARR29 pKa = 11.84SKK31 pKa = 10.88KK32 pKa = 9.0ILEE35 pKa = 4.24VTGISPDD42 pKa = 3.68LRR44 pKa = 11.84TDD46 pKa = 3.84DD47 pKa = 5.1LTDD50 pKa = 4.72DD51 pKa = 5.67DD52 pKa = 5.33IKK54 pKa = 10.99QLRR57 pKa = 11.84NALSEE62 pKa = 4.15YY63 pKa = 10.54NIEE66 pKa = 3.93GDD68 pKa = 3.53LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.58VTMNIKK78 pKa = 10.21RR79 pKa = 11.84LQEE82 pKa = 3.81INSYY86 pKa = 10.63RR87 pKa = 11.84GMRR90 pKa = 11.84HH91 pKa = 6.02KK92 pKa = 10.66RR93 pKa = 11.84GLPCRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 10.84TNARR107 pKa = 11.84TRR109 pKa = 11.84RR110 pKa = 11.84GKK112 pKa = 8.91KK113 pKa = 7.98TGPIAGKK120 pKa = 10.3KK121 pKa = 8.86KK122 pKa = 10.5
Molecular weight: 13.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
636768 |
31 |
2806 |
305.4 |
34.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.013 ± 0.049 | 1.244 ± 0.026 |
5.631 ± 0.041 | 7.053 ± 0.072 |
4.438 ± 0.048 | 6.107 ± 0.069 |
1.413 ± 0.019 | 8.366 ± 0.05 |
8.539 ± 0.068 | 9.047 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.515 ± 0.029 | 6.215 ± 0.079 |
3.22 ± 0.035 | 3.132 ± 0.035 |
3.536 ± 0.043 | 6.302 ± 0.063 |
5.406 ± 0.062 | 6.128 ± 0.041 |
0.671 ± 0.014 | 4.022 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
