
candidate division MSBL1 archaeon SCGC-AAA259M10
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Euryarchaeota incertae sedis; candidate division MSBL1
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
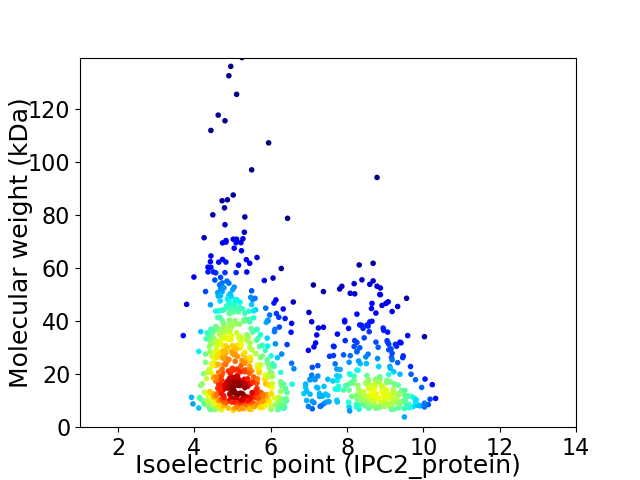
Virtual 2D-PAGE plot for 893 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A133UZA2|A0A133UZA2_9EURY Uncharacterized protein (Fragment) OS=candidate division MSBL1 archaeon SCGC-AAA259M10 OX=1698270 GN=AKJ40_02970 PE=4 SV=1
MM1 pKa = 7.12NRR3 pKa = 11.84SGTNKK8 pKa = 9.07TLLFLIITVTCFMVAASLPVVMGDD32 pKa = 3.99TIDD35 pKa = 4.59GEE37 pKa = 4.42ASVSNTSCEE46 pKa = 4.56FIDD49 pKa = 4.03KK50 pKa = 9.78TVDD53 pKa = 3.12TNTVDD58 pKa = 3.28PEE60 pKa = 4.5SYY62 pKa = 10.35FWIKK66 pKa = 8.37TQVRR70 pKa = 11.84DD71 pKa = 3.58SDD73 pKa = 3.95TLSDD77 pKa = 3.63IEE79 pKa = 4.23NVVVKK84 pKa = 10.81IFDD87 pKa = 3.43NQTAVGAQDD96 pKa = 3.65TKK98 pKa = 10.53RR99 pKa = 11.84SHH101 pKa = 4.15YY102 pKa = 8.61TFWFDD107 pKa = 5.43ANDD110 pKa = 3.62NSWNSNLNNTLGNPFIDD127 pKa = 4.08AAASEE132 pKa = 4.68YY133 pKa = 10.66PNDD136 pKa = 3.7ATVTEE141 pKa = 4.58DD142 pKa = 3.14NYY144 pKa = 11.42NFKK147 pKa = 10.54ILLNGTANPNQDD159 pKa = 2.4WNVYY163 pKa = 10.07VEE165 pKa = 4.53VNDD168 pKa = 3.92EE169 pKa = 4.22ASSEE173 pKa = 4.16NNGTHH178 pKa = 5.84TNALAVNTYY187 pKa = 10.3VNYY190 pKa = 10.07DD191 pKa = 3.38VKK193 pKa = 11.35ADD195 pKa = 4.44LIEE198 pKa = 4.15WTNLQPDD205 pKa = 4.22TNDD208 pKa = 3.51NPSDD212 pKa = 3.98SNPVIIEE219 pKa = 4.26NIEE222 pKa = 4.1TNVNYY227 pKa = 10.32DD228 pKa = 3.34VQTKK232 pKa = 10.75LSGDD236 pKa = 3.49WLNTNGDD243 pKa = 4.31TITVTEE249 pKa = 4.26TSFDD253 pKa = 3.73STDD256 pKa = 2.79SSGYY260 pKa = 9.18HH261 pKa = 5.47GQVSSSIFKK270 pKa = 10.43DD271 pKa = 3.44VYY273 pKa = 9.46TYY275 pKa = 10.99EE276 pKa = 4.44SYY278 pKa = 11.57GEE280 pKa = 4.45DD281 pKa = 3.06LTEE284 pKa = 4.64GINYY288 pKa = 9.45FLDD291 pKa = 3.96IPDD294 pKa = 4.12GTSSGTFTNTFEE306 pKa = 4.49IEE308 pKa = 4.56VVAHH312 pKa = 6.77
MM1 pKa = 7.12NRR3 pKa = 11.84SGTNKK8 pKa = 9.07TLLFLIITVTCFMVAASLPVVMGDD32 pKa = 3.99TIDD35 pKa = 4.59GEE37 pKa = 4.42ASVSNTSCEE46 pKa = 4.56FIDD49 pKa = 4.03KK50 pKa = 9.78TVDD53 pKa = 3.12TNTVDD58 pKa = 3.28PEE60 pKa = 4.5SYY62 pKa = 10.35FWIKK66 pKa = 8.37TQVRR70 pKa = 11.84DD71 pKa = 3.58SDD73 pKa = 3.95TLSDD77 pKa = 3.63IEE79 pKa = 4.23NVVVKK84 pKa = 10.81IFDD87 pKa = 3.43NQTAVGAQDD96 pKa = 3.65TKK98 pKa = 10.53RR99 pKa = 11.84SHH101 pKa = 4.15YY102 pKa = 8.61TFWFDD107 pKa = 5.43ANDD110 pKa = 3.62NSWNSNLNNTLGNPFIDD127 pKa = 4.08AAASEE132 pKa = 4.68YY133 pKa = 10.66PNDD136 pKa = 3.7ATVTEE141 pKa = 4.58DD142 pKa = 3.14NYY144 pKa = 11.42NFKK147 pKa = 10.54ILLNGTANPNQDD159 pKa = 2.4WNVYY163 pKa = 10.07VEE165 pKa = 4.53VNDD168 pKa = 3.92EE169 pKa = 4.22ASSEE173 pKa = 4.16NNGTHH178 pKa = 5.84TNALAVNTYY187 pKa = 10.3VNYY190 pKa = 10.07DD191 pKa = 3.38VKK193 pKa = 11.35ADD195 pKa = 4.44LIEE198 pKa = 4.15WTNLQPDD205 pKa = 4.22TNDD208 pKa = 3.51NPSDD212 pKa = 3.98SNPVIIEE219 pKa = 4.26NIEE222 pKa = 4.1TNVNYY227 pKa = 10.32DD228 pKa = 3.34VQTKK232 pKa = 10.75LSGDD236 pKa = 3.49WLNTNGDD243 pKa = 4.31TITVTEE249 pKa = 4.26TSFDD253 pKa = 3.73STDD256 pKa = 2.79SSGYY260 pKa = 9.18HH261 pKa = 5.47GQVSSSIFKK270 pKa = 10.43DD271 pKa = 3.44VYY273 pKa = 9.46TYY275 pKa = 10.99EE276 pKa = 4.44SYY278 pKa = 11.57GEE280 pKa = 4.45DD281 pKa = 3.06LTEE284 pKa = 4.64GINYY288 pKa = 9.45FLDD291 pKa = 3.96IPDD294 pKa = 4.12GTSSGTFTNTFEE306 pKa = 4.49IEE308 pKa = 4.56VVAHH312 pKa = 6.77
Molecular weight: 34.58 kDa
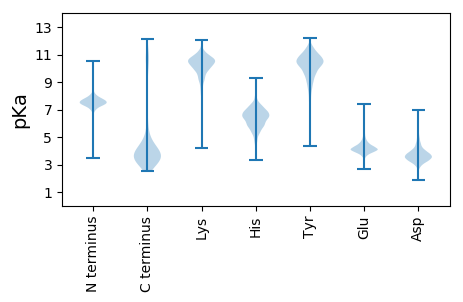
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133UYJ2|A0A133UYJ2_9EURY 4Fe-4S ferredoxin (Fragment) OS=candidate division MSBL1 archaeon SCGC-AAA259M10 OX=1698270 GN=AKJ40_03640 PE=4 SV=1
MM1 pKa = 7.39RR2 pKa = 11.84RR3 pKa = 11.84EE4 pKa = 4.24GSWAWADD11 pKa = 3.45MLLDD15 pKa = 5.29FIEE18 pKa = 5.44NPQKK22 pKa = 10.3WLRR25 pKa = 11.84EE26 pKa = 3.79CHH28 pKa = 5.59TRR30 pKa = 11.84SNVEE34 pKa = 4.05SGFSTFKK41 pKa = 10.72RR42 pKa = 11.84HH43 pKa = 5.87FLSPLRR49 pKa = 11.84KK50 pKa = 9.5CIQRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 7.84TEE59 pKa = 3.53AFARR63 pKa = 11.84ACDD66 pKa = 3.98YY67 pKa = 10.69NLKK70 pKa = 9.63RR71 pKa = 11.84ASYY74 pKa = 10.09VRR76 pKa = 11.84RR77 pKa = 11.84QEE79 pKa = 3.98GLTAA83 pKa = 4.79
MM1 pKa = 7.39RR2 pKa = 11.84RR3 pKa = 11.84EE4 pKa = 4.24GSWAWADD11 pKa = 3.45MLLDD15 pKa = 5.29FIEE18 pKa = 5.44NPQKK22 pKa = 10.3WLRR25 pKa = 11.84EE26 pKa = 3.79CHH28 pKa = 5.59TRR30 pKa = 11.84SNVEE34 pKa = 4.05SGFSTFKK41 pKa = 10.72RR42 pKa = 11.84HH43 pKa = 5.87FLSPLRR49 pKa = 11.84KK50 pKa = 9.5CIQRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 7.84TEE59 pKa = 3.53AFARR63 pKa = 11.84ACDD66 pKa = 3.98YY67 pKa = 10.69NLKK70 pKa = 9.63RR71 pKa = 11.84ASYY74 pKa = 10.09VRR76 pKa = 11.84RR77 pKa = 11.84QEE79 pKa = 3.98GLTAA83 pKa = 4.79
Molecular weight: 10.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
200716 |
33 |
1234 |
224.8 |
25.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.858 ± 0.082 | 1.003 ± 0.034 |
5.787 ± 0.064 | 10.172 ± 0.123 |
4.036 ± 0.072 | 7.458 ± 0.085 |
1.693 ± 0.039 | 7.013 ± 0.08 |
7.561 ± 0.098 | 9.254 ± 0.095 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.198 ± 0.036 | 3.747 ± 0.052 |
4.05 ± 0.053 | 2.206 ± 0.042 |
5.755 ± 0.081 | 6.382 ± 0.065 |
4.823 ± 0.068 | 6.878 ± 0.061 |
1.098 ± 0.034 | 3.029 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
