
Sugarcane yellow leaf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
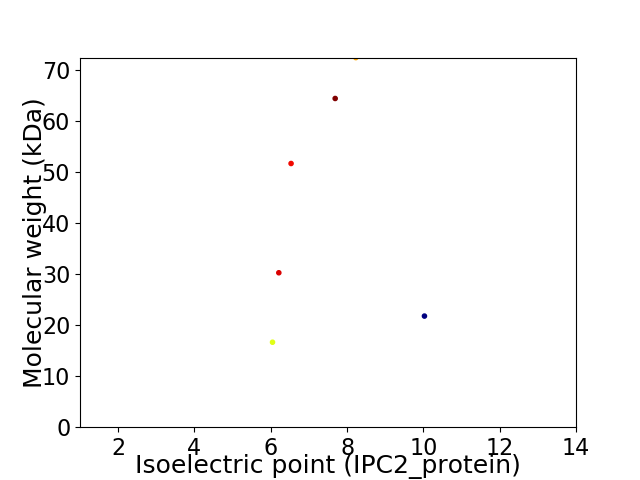
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
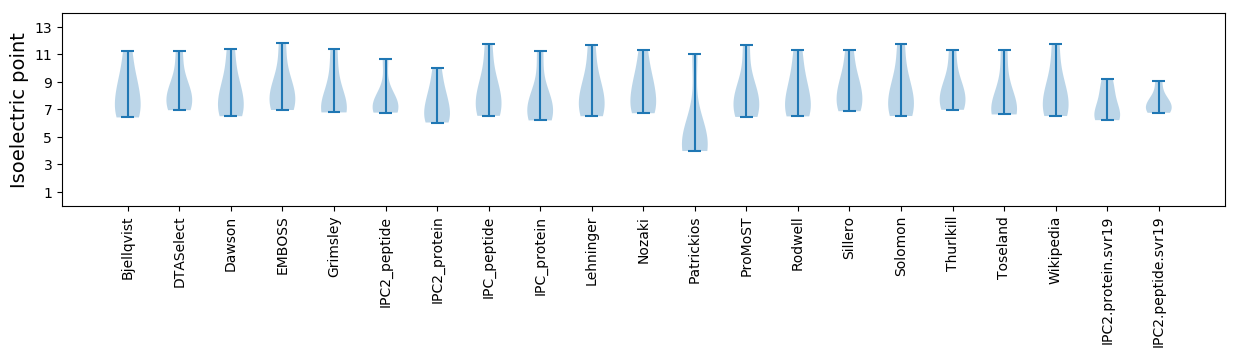
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9WI47|Q9WI47_9LUTE Movement protein OS=Sugarcane yellow leaf virus OX=94290 GN=ORF4 PE=2 SV=1
MM1 pKa = 7.91SEE3 pKa = 4.0DD4 pKa = 3.35ALTVVDD10 pKa = 6.27RR11 pKa = 11.84LGQWSWSGLPQDD23 pKa = 4.29LDD25 pKa = 3.7EE26 pKa = 5.86YY27 pKa = 11.52DD28 pKa = 4.31DD29 pKa = 4.13VEE31 pKa = 4.47HH32 pKa = 7.27VLEE35 pKa = 4.33EE36 pKa = 4.28TLCEE40 pKa = 4.08DD41 pKa = 4.33RR42 pKa = 11.84EE43 pKa = 4.2EE44 pKa = 4.33EE45 pKa = 4.08ATGMFSLSRR54 pKa = 11.84LTISKK59 pKa = 7.72PTQPGSSNSDD69 pKa = 2.74RR70 pKa = 11.84TYY72 pKa = 11.49LSTQRR77 pKa = 11.84STMAYY82 pKa = 9.43SKK84 pKa = 9.19PTMSIKK90 pKa = 10.6SQVSLFSITHH100 pKa = 6.69APPTQLQVQSHH111 pKa = 6.52LKK113 pKa = 9.14WIHH116 pKa = 6.27PAPKK120 pKa = 9.34QQQAPRR126 pKa = 11.84LLASPSRR133 pKa = 11.84GTPRR137 pKa = 11.84KK138 pKa = 9.38SSRR141 pKa = 11.84PPSSGGKK148 pKa = 9.51ISS150 pKa = 3.16
MM1 pKa = 7.91SEE3 pKa = 4.0DD4 pKa = 3.35ALTVVDD10 pKa = 6.27RR11 pKa = 11.84LGQWSWSGLPQDD23 pKa = 4.29LDD25 pKa = 3.7EE26 pKa = 5.86YY27 pKa = 11.52DD28 pKa = 4.31DD29 pKa = 4.13VEE31 pKa = 4.47HH32 pKa = 7.27VLEE35 pKa = 4.33EE36 pKa = 4.28TLCEE40 pKa = 4.08DD41 pKa = 4.33RR42 pKa = 11.84EE43 pKa = 4.2EE44 pKa = 4.33EE45 pKa = 4.08ATGMFSLSRR54 pKa = 11.84LTISKK59 pKa = 7.72PTQPGSSNSDD69 pKa = 2.74RR70 pKa = 11.84TYY72 pKa = 11.49LSTQRR77 pKa = 11.84STMAYY82 pKa = 9.43SKK84 pKa = 9.19PTMSIKK90 pKa = 10.6SQVSLFSITHH100 pKa = 6.69APPTQLQVQSHH111 pKa = 6.52LKK113 pKa = 9.14WIHH116 pKa = 6.27PAPKK120 pKa = 9.34QQQAPRR126 pKa = 11.84LLASPSRR133 pKa = 11.84GTPRR137 pKa = 11.84KK138 pKa = 9.38SSRR141 pKa = 11.84PPSSGGKK148 pKa = 9.51ISS150 pKa = 3.16
Molecular weight: 16.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QQN8|Q9QQN8_9LUTE RNA-directed RNA polymerase (Fragment) OS=Sugarcane yellow leaf virus OX=94290 PE=4 SV=1
MM1 pKa = 6.87NTGANRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84NVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ANRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84QTRR23 pKa = 11.84PVVVVRR29 pKa = 11.84APPGPRR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84ARR43 pKa = 11.84VGGNAVRR50 pKa = 11.84GPGGRR55 pKa = 11.84SNRR58 pKa = 11.84DD59 pKa = 3.02VLTFTVDD66 pKa = 3.38DD67 pKa = 4.74LKK69 pKa = 11.69ANSTGILKK77 pKa = 10.11FGPNLSQYY85 pKa = 11.29AAFNNGLLKK94 pKa = 10.37AYY96 pKa = 9.79HH97 pKa = 6.59EE98 pKa = 4.35YY99 pKa = 10.72KK100 pKa = 9.38ITSLTIQYY108 pKa = 9.67NSCSSDD114 pKa = 3.1ATPGAIALEE123 pKa = 4.33VDD125 pKa = 4.38TSCSQTTTGSKK136 pKa = 8.38ITSFPVKK143 pKa = 10.24RR144 pKa = 11.84NAKK147 pKa = 9.58KK148 pKa = 10.49VFPAPFIRR156 pKa = 11.84GKK158 pKa = 10.79DD159 pKa = 3.78FMTTSADD166 pKa = 4.07QFWLLYY172 pKa = 9.95KK173 pKa = 11.05GNGDD177 pKa = 3.49SSLAGQFVCRR187 pKa = 11.84FEE189 pKa = 6.15CLFQNPKK196 pKa = 10.36
MM1 pKa = 6.87NTGANRR7 pKa = 11.84SRR9 pKa = 11.84RR10 pKa = 11.84NVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ANRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84QTRR23 pKa = 11.84PVVVVRR29 pKa = 11.84APPGPRR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84ARR43 pKa = 11.84VGGNAVRR50 pKa = 11.84GPGGRR55 pKa = 11.84SNRR58 pKa = 11.84DD59 pKa = 3.02VLTFTVDD66 pKa = 3.38DD67 pKa = 4.74LKK69 pKa = 11.69ANSTGILKK77 pKa = 10.11FGPNLSQYY85 pKa = 11.29AAFNNGLLKK94 pKa = 10.37AYY96 pKa = 9.79HH97 pKa = 6.59EE98 pKa = 4.35YY99 pKa = 10.72KK100 pKa = 9.38ITSLTIQYY108 pKa = 9.67NSCSSDD114 pKa = 3.1ATPGAIALEE123 pKa = 4.33VDD125 pKa = 4.38TSCSQTTTGSKK136 pKa = 8.38ITSFPVKK143 pKa = 10.24RR144 pKa = 11.84NAKK147 pKa = 9.58KK148 pKa = 10.49VFPAPFIRR156 pKa = 11.84GKK158 pKa = 10.79DD159 pKa = 3.78FMTTSADD166 pKa = 4.07QFWLLYY172 pKa = 9.95KK173 pKa = 11.05GNGDD177 pKa = 3.49SSLAGQFVCRR187 pKa = 11.84FEE189 pKa = 6.15CLFQNPKK196 pKa = 10.36
Molecular weight: 21.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2303 |
150 |
650 |
383.8 |
42.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.121 ± 0.799 | 1.433 ± 0.282 |
4.733 ± 0.247 | 5.688 ± 0.669 |
3.821 ± 0.407 | 6.166 ± 0.504 |
2.301 ± 0.429 | 3.865 ± 0.485 |
5.732 ± 0.572 | 8.684 ± 0.677 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.911 ± 0.358 | 4.472 ± 0.418 |
7.338 ± 1.088 | 4.038 ± 0.364 |
6.34 ± 1.004 | 8.988 ± 0.869 |
6.036 ± 0.576 | 6.426 ± 0.29 |
1.824 ± 0.304 | 3.04 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
