
Micromonospora sp. MH33
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Micromonospora; unclassified Micromonospora
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
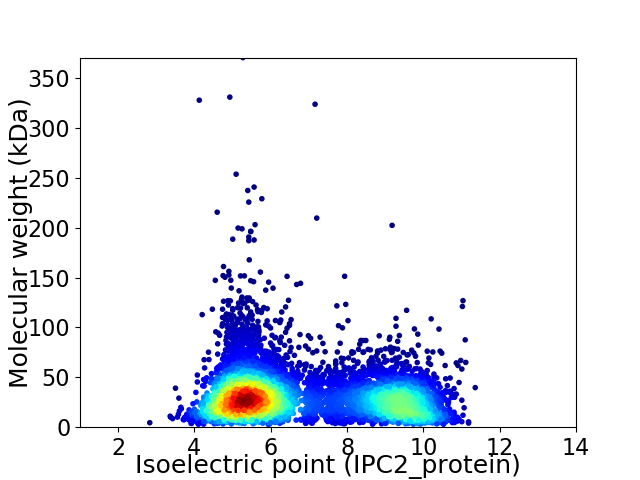
Virtual 2D-PAGE plot for 7025 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P8AQA1|A0A2P8AQA1_9ACTN NTP_transf_2 domain-containing protein OS=Micromonospora sp. MH33 OX=1945509 GN=B0E53_05434 PE=4 SV=1
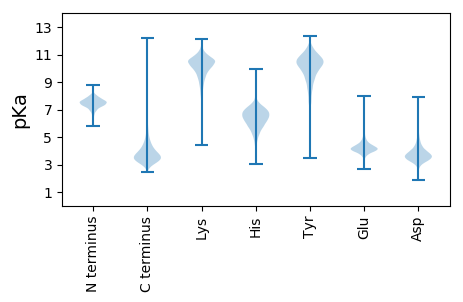
MM1 pKa = 8.19DD2 pKa = 3.7SHH4 pKa = 5.86QTLHH8 pKa = 7.34DD9 pKa = 3.84FVLNLLTDD17 pKa = 4.23PDD19 pKa = 3.68ARR21 pKa = 11.84SAFDD25 pKa = 4.84LDD27 pKa = 4.23PEE29 pKa = 4.73GALQAAGLTDD39 pKa = 3.29VTAADD44 pKa = 3.9VQDD47 pKa = 3.98VVPLVVDD54 pKa = 3.87YY55 pKa = 11.4APGAAGLAPVAPVAGQLGLDD75 pKa = 4.35PLTSDD80 pKa = 3.06ATDD83 pKa = 3.21VVGQLQAVPHH93 pKa = 5.8QISVNSSHH101 pKa = 7.0SGVDD105 pKa = 3.21MSAGALGAIAVDD117 pKa = 3.7PAGLYY122 pKa = 10.49AGAALVPGIGLGIGPGGLGTDD143 pKa = 3.46LTGVHH148 pKa = 6.93DD149 pKa = 4.3VANTLDD155 pKa = 4.05ADD157 pKa = 3.99VVGAVDD163 pKa = 3.55NAADD167 pKa = 3.87PVVGDD172 pKa = 4.0LAGTAGDD179 pKa = 4.27PLGTTAGVDD188 pKa = 3.69LLHH191 pKa = 7.45DD192 pKa = 4.44GGVLGGDD199 pKa = 3.69LGVLSGTQAQVDD211 pKa = 4.36GVVGSLGVHH220 pKa = 4.33QTLGGFGLGDD230 pKa = 3.52GSGVVPPVHH239 pKa = 6.99APATVDD245 pKa = 3.59GVTHH249 pKa = 5.83QVDD252 pKa = 3.56STLAGVTGTVSGVTGTVGGLTGGVTDD278 pKa = 5.38GVTGDD283 pKa = 4.12AGVHH287 pKa = 5.3GAGVHH292 pKa = 5.99GDD294 pKa = 3.5VHH296 pKa = 6.91GSTDD300 pKa = 3.23GGLLGGLLL308 pKa = 3.81
MM1 pKa = 8.19DD2 pKa = 3.7SHH4 pKa = 5.86QTLHH8 pKa = 7.34DD9 pKa = 3.84FVLNLLTDD17 pKa = 4.23PDD19 pKa = 3.68ARR21 pKa = 11.84SAFDD25 pKa = 4.84LDD27 pKa = 4.23PEE29 pKa = 4.73GALQAAGLTDD39 pKa = 3.29VTAADD44 pKa = 3.9VQDD47 pKa = 3.98VVPLVVDD54 pKa = 3.87YY55 pKa = 11.4APGAAGLAPVAPVAGQLGLDD75 pKa = 4.35PLTSDD80 pKa = 3.06ATDD83 pKa = 3.21VVGQLQAVPHH93 pKa = 5.8QISVNSSHH101 pKa = 7.0SGVDD105 pKa = 3.21MSAGALGAIAVDD117 pKa = 3.7PAGLYY122 pKa = 10.49AGAALVPGIGLGIGPGGLGTDD143 pKa = 3.46LTGVHH148 pKa = 6.93DD149 pKa = 4.3VANTLDD155 pKa = 4.05ADD157 pKa = 3.99VVGAVDD163 pKa = 3.55NAADD167 pKa = 3.87PVVGDD172 pKa = 4.0LAGTAGDD179 pKa = 4.27PLGTTAGVDD188 pKa = 3.69LLHH191 pKa = 7.45DD192 pKa = 4.44GGVLGGDD199 pKa = 3.69LGVLSGTQAQVDD211 pKa = 4.36GVVGSLGVHH220 pKa = 4.33QTLGGFGLGDD230 pKa = 3.52GSGVVPPVHH239 pKa = 6.99APATVDD245 pKa = 3.59GVTHH249 pKa = 5.83QVDD252 pKa = 3.56STLAGVTGTVSGVTGTVGGLTGGVTDD278 pKa = 5.38GVTGDD283 pKa = 4.12AGVHH287 pKa = 5.3GAGVHH292 pKa = 5.99GDD294 pKa = 3.5VHH296 pKa = 6.91GSTDD300 pKa = 3.23GGLLGGLLL308 pKa = 3.81
Molecular weight: 29.16 kDa
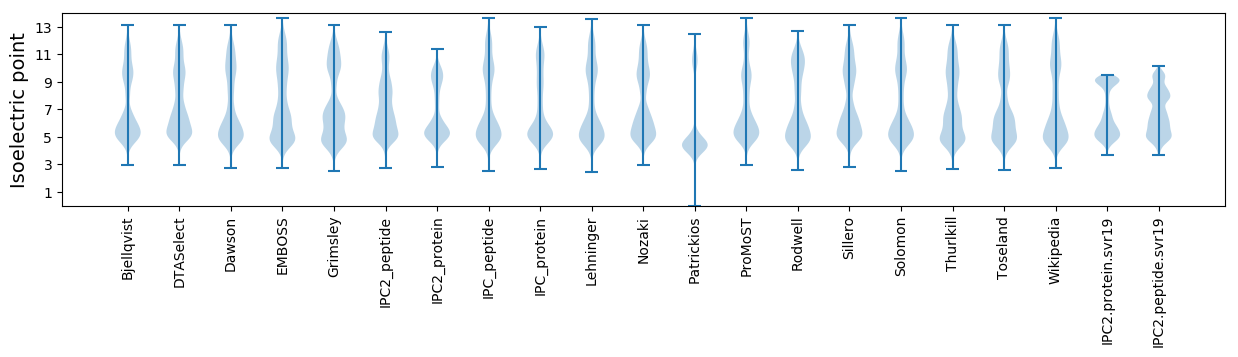
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P8ALI0|A0A2P8ALI0_9ACTN Uncharacterized protein OS=Micromonospora sp. MH33 OX=1945509 GN=B0E53_06804 PE=4 SV=1
MM1 pKa = 7.33GGVRR5 pKa = 11.84TGRR8 pKa = 11.84HH9 pKa = 5.02RR10 pKa = 11.84PAHH13 pKa = 6.83RR14 pKa = 11.84PRR16 pKa = 11.84QQHH19 pKa = 4.82RR20 pKa = 11.84RR21 pKa = 11.84LRR23 pKa = 11.84RR24 pKa = 11.84PDD26 pKa = 3.35LPRR29 pKa = 11.84LRR31 pKa = 11.84GQGARR36 pKa = 11.84HH37 pKa = 5.84LRR39 pKa = 11.84RR40 pKa = 11.84ARR42 pKa = 11.84RR43 pKa = 11.84VPGQRR48 pKa = 11.84QRR50 pKa = 11.84RR51 pKa = 11.84QRR53 pKa = 11.84RR54 pKa = 11.84LRR56 pKa = 11.84PHH58 pKa = 7.38LLPVRR63 pKa = 11.84PRR65 pKa = 11.84RR66 pKa = 11.84PGRR69 pKa = 11.84HH70 pKa = 4.38RR71 pKa = 11.84RR72 pKa = 11.84HH73 pKa = 5.85RR74 pKa = 11.84VLLVAGGPAPGRR86 pKa = 11.84PGVAPGGVRR95 pKa = 11.84ARR97 pKa = 11.84PRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84QRR104 pKa = 11.84HH105 pKa = 3.56VHH107 pKa = 5.63PRR109 pKa = 11.84HH110 pKa = 5.67ARR112 pKa = 11.84RR113 pKa = 11.84VRR115 pKa = 11.84PPARR119 pKa = 11.84PVTRR123 pKa = 11.84RR124 pKa = 11.84PVQGVRR130 pKa = 11.84RR131 pKa = 11.84GRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84HH136 pKa = 5.01RR137 pKa = 11.84LRR139 pKa = 11.84RR140 pKa = 11.84GRR142 pKa = 11.84RR143 pKa = 11.84HH144 pKa = 5.77AAAGTPLRR152 pKa = 11.84RR153 pKa = 11.84PPQRR157 pKa = 11.84PPDD160 pKa = 3.62PRR162 pKa = 11.84RR163 pKa = 11.84AARR166 pKa = 11.84QRR168 pKa = 11.84GQPGRR173 pKa = 11.84RR174 pKa = 11.84QQRR177 pKa = 11.84PHH179 pKa = 6.81RR180 pKa = 11.84PQRR183 pKa = 11.84PRR185 pKa = 11.84PAAGHH190 pKa = 6.14PPGPRR195 pKa = 11.84ARR197 pKa = 11.84AAVPVRR203 pKa = 11.84RR204 pKa = 11.84GRR206 pKa = 11.84GRR208 pKa = 11.84GPRR211 pKa = 11.84HH212 pKa = 4.88RR213 pKa = 11.84HH214 pKa = 4.57RR215 pKa = 11.84PRR217 pKa = 11.84RR218 pKa = 11.84PHH220 pKa = 6.58RR221 pKa = 11.84GAGAAGHH228 pKa = 6.69LRR230 pKa = 11.84AGPPRR235 pKa = 11.84RR236 pKa = 11.84PAAAARR242 pKa = 11.84LDD244 pKa = 4.03QVEE247 pKa = 4.16HH248 pKa = 6.86RR249 pKa = 11.84PRR251 pKa = 11.84PGRR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84GDD261 pKa = 3.01QDD263 pKa = 3.59GPRR266 pKa = 11.84HH267 pKa = 5.85AARR270 pKa = 11.84PAPGHH275 pKa = 5.49PARR278 pKa = 11.84RR279 pKa = 11.84PAVPAHH285 pKa = 6.11RR286 pKa = 11.84LDD288 pKa = 3.49RR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84GRR293 pKa = 11.84PARR296 pKa = 11.84RR297 pKa = 11.84RR298 pKa = 11.84PALARR303 pKa = 11.84RR304 pKa = 11.84GPAPPRR310 pKa = 11.84RR311 pKa = 11.84RR312 pKa = 11.84LLVRR316 pKa = 11.84HH317 pKa = 5.56QRR319 pKa = 11.84HH320 pKa = 4.0QRR322 pKa = 11.84ARR324 pKa = 11.84HH325 pKa = 5.64PGAGPRR331 pKa = 11.84AGRR334 pKa = 11.84RR335 pKa = 11.84AGRR338 pKa = 11.84AGPGPRR344 pKa = 3.94
MM1 pKa = 7.33GGVRR5 pKa = 11.84TGRR8 pKa = 11.84HH9 pKa = 5.02RR10 pKa = 11.84PAHH13 pKa = 6.83RR14 pKa = 11.84PRR16 pKa = 11.84QQHH19 pKa = 4.82RR20 pKa = 11.84RR21 pKa = 11.84LRR23 pKa = 11.84RR24 pKa = 11.84PDD26 pKa = 3.35LPRR29 pKa = 11.84LRR31 pKa = 11.84GQGARR36 pKa = 11.84HH37 pKa = 5.84LRR39 pKa = 11.84RR40 pKa = 11.84ARR42 pKa = 11.84RR43 pKa = 11.84VPGQRR48 pKa = 11.84QRR50 pKa = 11.84RR51 pKa = 11.84QRR53 pKa = 11.84RR54 pKa = 11.84LRR56 pKa = 11.84PHH58 pKa = 7.38LLPVRR63 pKa = 11.84PRR65 pKa = 11.84RR66 pKa = 11.84PGRR69 pKa = 11.84HH70 pKa = 4.38RR71 pKa = 11.84RR72 pKa = 11.84HH73 pKa = 5.85RR74 pKa = 11.84VLLVAGGPAPGRR86 pKa = 11.84PGVAPGGVRR95 pKa = 11.84ARR97 pKa = 11.84PRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84QRR104 pKa = 11.84HH105 pKa = 3.56VHH107 pKa = 5.63PRR109 pKa = 11.84HH110 pKa = 5.67ARR112 pKa = 11.84RR113 pKa = 11.84VRR115 pKa = 11.84PPARR119 pKa = 11.84PVTRR123 pKa = 11.84RR124 pKa = 11.84PVQGVRR130 pKa = 11.84RR131 pKa = 11.84GRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84HH136 pKa = 5.01RR137 pKa = 11.84LRR139 pKa = 11.84RR140 pKa = 11.84GRR142 pKa = 11.84RR143 pKa = 11.84HH144 pKa = 5.77AAAGTPLRR152 pKa = 11.84RR153 pKa = 11.84PPQRR157 pKa = 11.84PPDD160 pKa = 3.62PRR162 pKa = 11.84RR163 pKa = 11.84AARR166 pKa = 11.84QRR168 pKa = 11.84GQPGRR173 pKa = 11.84RR174 pKa = 11.84QQRR177 pKa = 11.84PHH179 pKa = 6.81RR180 pKa = 11.84PQRR183 pKa = 11.84PRR185 pKa = 11.84PAAGHH190 pKa = 6.14PPGPRR195 pKa = 11.84ARR197 pKa = 11.84AAVPVRR203 pKa = 11.84RR204 pKa = 11.84GRR206 pKa = 11.84GRR208 pKa = 11.84GPRR211 pKa = 11.84HH212 pKa = 4.88RR213 pKa = 11.84HH214 pKa = 4.57RR215 pKa = 11.84PRR217 pKa = 11.84RR218 pKa = 11.84PHH220 pKa = 6.58RR221 pKa = 11.84GAGAAGHH228 pKa = 6.69LRR230 pKa = 11.84AGPPRR235 pKa = 11.84RR236 pKa = 11.84PAAAARR242 pKa = 11.84LDD244 pKa = 4.03QVEE247 pKa = 4.16HH248 pKa = 6.86RR249 pKa = 11.84PRR251 pKa = 11.84PGRR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84GDD261 pKa = 3.01QDD263 pKa = 3.59GPRR266 pKa = 11.84HH267 pKa = 5.85AARR270 pKa = 11.84PAPGHH275 pKa = 5.49PARR278 pKa = 11.84RR279 pKa = 11.84PAVPAHH285 pKa = 6.11RR286 pKa = 11.84LDD288 pKa = 3.49RR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84GRR293 pKa = 11.84PARR296 pKa = 11.84RR297 pKa = 11.84RR298 pKa = 11.84PALARR303 pKa = 11.84RR304 pKa = 11.84GPAPPRR310 pKa = 11.84RR311 pKa = 11.84RR312 pKa = 11.84LLVRR316 pKa = 11.84HH317 pKa = 5.56QRR319 pKa = 11.84HH320 pKa = 4.0QRR322 pKa = 11.84ARR324 pKa = 11.84HH325 pKa = 5.64PGAGPRR331 pKa = 11.84AGRR334 pKa = 11.84RR335 pKa = 11.84AGRR338 pKa = 11.84AGPGPRR344 pKa = 3.94
Molecular weight: 39.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2123840 |
29 |
3432 |
302.3 |
32.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.954 ± 0.049 | 0.785 ± 0.009 |
5.983 ± 0.022 | 5.016 ± 0.03 |
2.63 ± 0.018 | 9.474 ± 0.035 |
2.243 ± 0.019 | 3.054 ± 0.021 |
1.714 ± 0.025 | 10.416 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.611 ± 0.012 | 1.78 ± 0.019 |
6.485 ± 0.031 | 2.749 ± 0.023 |
8.708 ± 0.05 | 4.705 ± 0.027 |
6.042 ± 0.029 | 8.989 ± 0.033 |
1.614 ± 0.014 | 2.047 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
