
Planctomycetes bacterium Pla163
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; unclassified Planctomycetes
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
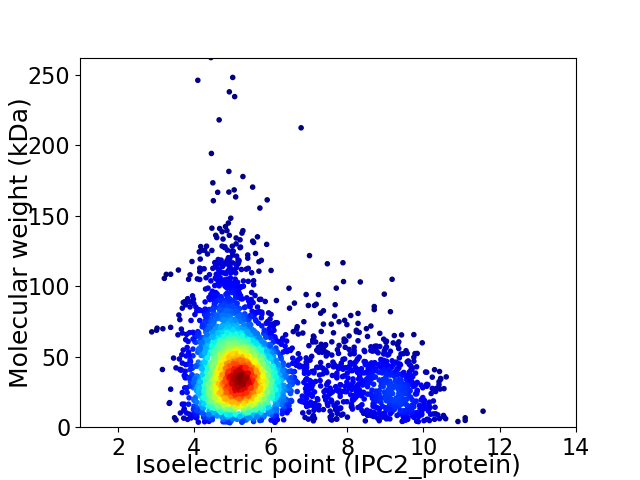
Virtual 2D-PAGE plot for 3741 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
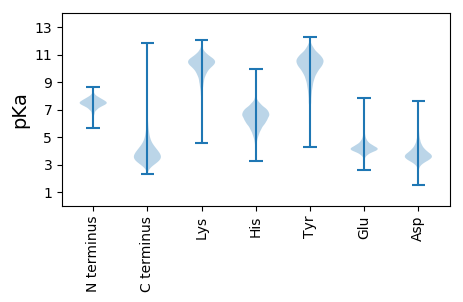
Protein with the lowest isoelectric point:
>tr|A0A518D480|A0A518D480_9BACT Phosphatidylglycerophosphatase A OS=Planctomycetes bacterium Pla163 OX=2528008 GN=pgpA PE=4 SV=1
MM1 pKa = 7.43NPLLSGALFTALSPLALAGGTEE23 pKa = 4.61ASSDD27 pKa = 3.12WLGLDD32 pKa = 3.47EE33 pKa = 5.07EE34 pKa = 4.98LNAVASNVSLQGGGVEE50 pKa = 4.71LGALFRR56 pKa = 11.84GAYY59 pKa = 9.63VMADD63 pKa = 2.95GTDD66 pKa = 3.84FNGFADD72 pKa = 4.35GNGDD76 pKa = 4.49GNPDD80 pKa = 3.63DD81 pKa = 3.9QLGMVVDD88 pKa = 5.22DD89 pKa = 5.03AEE91 pKa = 4.51LWSQGTVGDD100 pKa = 4.12YY101 pKa = 9.84FWRR104 pKa = 11.84ISVDD108 pKa = 3.61FGTQFSPYY116 pKa = 9.83FIGGLIGTGAGTSAFANTARR136 pKa = 11.84TDD138 pKa = 3.77TLEE141 pKa = 4.35DD142 pKa = 4.05AYY144 pKa = 10.18IDD146 pKa = 3.58WRR148 pKa = 11.84FSDD151 pKa = 3.81QFSLRR156 pKa = 11.84LGHH159 pKa = 7.04FILPTTFSAASNPDD173 pKa = 3.35SLLMLGRR180 pKa = 11.84TTSGEE185 pKa = 3.96YY186 pKa = 9.42FHH188 pKa = 7.64NYY190 pKa = 9.82DD191 pKa = 4.25LGAMLSGDD199 pKa = 3.46YY200 pKa = 10.78EE201 pKa = 4.09QFQWHH206 pKa = 6.39LAATNGFDD214 pKa = 4.03GTGDD218 pKa = 3.53EE219 pKa = 3.97QRR221 pKa = 11.84LFGRR225 pKa = 11.84AIYY228 pKa = 10.56NLGGGQNATEE238 pKa = 4.34GALGADD244 pKa = 4.0EE245 pKa = 4.95NLNAAIGATFWDD257 pKa = 5.2DD258 pKa = 3.56GTVDD262 pKa = 4.06DD263 pKa = 4.84SDD265 pKa = 4.21VVAFDD270 pKa = 3.35VSATVGQISFSGEE283 pKa = 3.51FVTYY287 pKa = 10.72GDD289 pKa = 3.61AHH291 pKa = 5.9ATAAAGTFAGATNLGYY307 pKa = 8.44TAGSGPSVWGLTVSYY322 pKa = 10.45LLNEE326 pKa = 4.19EE327 pKa = 4.1WEE329 pKa = 4.15IAARR333 pKa = 11.84MEE335 pKa = 5.58DD336 pKa = 3.78MDD338 pKa = 5.49DD339 pKa = 3.7LANTEE344 pKa = 3.97IMSFGLNYY352 pKa = 10.13YY353 pKa = 9.97QLGHH357 pKa = 5.44NAKK360 pKa = 8.22WQLMLMSVSSDD371 pKa = 3.34VAVNEE376 pKa = 4.19GSVIALGVTVGSNGG390 pKa = 3.16
MM1 pKa = 7.43NPLLSGALFTALSPLALAGGTEE23 pKa = 4.61ASSDD27 pKa = 3.12WLGLDD32 pKa = 3.47EE33 pKa = 5.07EE34 pKa = 4.98LNAVASNVSLQGGGVEE50 pKa = 4.71LGALFRR56 pKa = 11.84GAYY59 pKa = 9.63VMADD63 pKa = 2.95GTDD66 pKa = 3.84FNGFADD72 pKa = 4.35GNGDD76 pKa = 4.49GNPDD80 pKa = 3.63DD81 pKa = 3.9QLGMVVDD88 pKa = 5.22DD89 pKa = 5.03AEE91 pKa = 4.51LWSQGTVGDD100 pKa = 4.12YY101 pKa = 9.84FWRR104 pKa = 11.84ISVDD108 pKa = 3.61FGTQFSPYY116 pKa = 9.83FIGGLIGTGAGTSAFANTARR136 pKa = 11.84TDD138 pKa = 3.77TLEE141 pKa = 4.35DD142 pKa = 4.05AYY144 pKa = 10.18IDD146 pKa = 3.58WRR148 pKa = 11.84FSDD151 pKa = 3.81QFSLRR156 pKa = 11.84LGHH159 pKa = 7.04FILPTTFSAASNPDD173 pKa = 3.35SLLMLGRR180 pKa = 11.84TTSGEE185 pKa = 3.96YY186 pKa = 9.42FHH188 pKa = 7.64NYY190 pKa = 9.82DD191 pKa = 4.25LGAMLSGDD199 pKa = 3.46YY200 pKa = 10.78EE201 pKa = 4.09QFQWHH206 pKa = 6.39LAATNGFDD214 pKa = 4.03GTGDD218 pKa = 3.53EE219 pKa = 3.97QRR221 pKa = 11.84LFGRR225 pKa = 11.84AIYY228 pKa = 10.56NLGGGQNATEE238 pKa = 4.34GALGADD244 pKa = 4.0EE245 pKa = 4.95NLNAAIGATFWDD257 pKa = 5.2DD258 pKa = 3.56GTVDD262 pKa = 4.06DD263 pKa = 4.84SDD265 pKa = 4.21VVAFDD270 pKa = 3.35VSATVGQISFSGEE283 pKa = 3.51FVTYY287 pKa = 10.72GDD289 pKa = 3.61AHH291 pKa = 5.9ATAAAGTFAGATNLGYY307 pKa = 8.44TAGSGPSVWGLTVSYY322 pKa = 10.45LLNEE326 pKa = 4.19EE327 pKa = 4.1WEE329 pKa = 4.15IAARR333 pKa = 11.84MEE335 pKa = 5.58DD336 pKa = 3.78MDD338 pKa = 5.49DD339 pKa = 3.7LANTEE344 pKa = 3.97IMSFGLNYY352 pKa = 10.13YY353 pKa = 9.97QLGHH357 pKa = 5.44NAKK360 pKa = 8.22WQLMLMSVSSDD371 pKa = 3.34VAVNEE376 pKa = 4.19GSVIALGVTVGSNGG390 pKa = 3.16
Molecular weight: 41.03 kDa
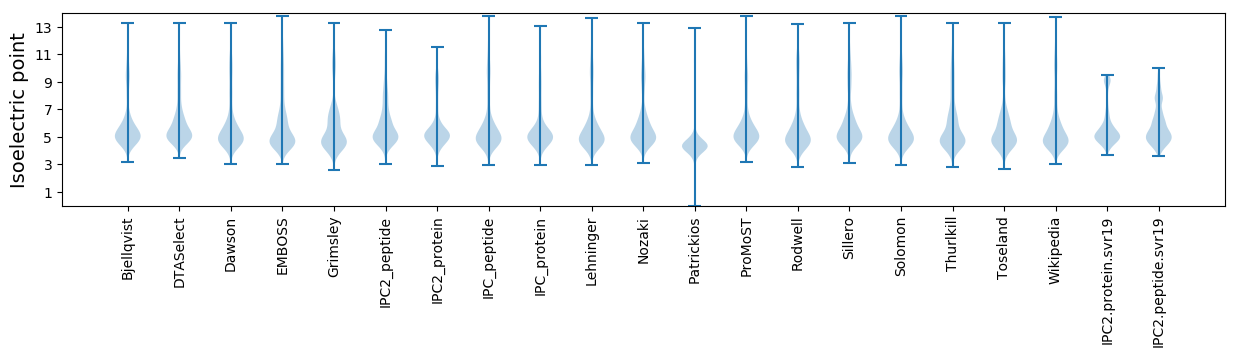
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518D0A3|A0A518D0A3_9BACT Uncharacterized protein OS=Planctomycetes bacterium Pla163 OX=2528008 GN=Pla163_19930 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.6SKK4 pKa = 10.95GSVKK8 pKa = 10.19KK9 pKa = 10.61RR10 pKa = 11.84FRR12 pKa = 11.84TTKK15 pKa = 10.0NGKK18 pKa = 9.75LIANKK23 pKa = 7.23TARR26 pKa = 11.84GHH28 pKa = 4.21MHH30 pKa = 6.63APKK33 pKa = 10.15NGKK36 pKa = 8.69ARR38 pKa = 11.84RR39 pKa = 11.84GKK41 pKa = 10.22RR42 pKa = 11.84KK43 pKa = 8.98PLVISGTWAVLMKK56 pKa = 10.94RR57 pKa = 11.84MMM59 pKa = 4.28
MM1 pKa = 7.46KK2 pKa = 10.6SKK4 pKa = 10.95GSVKK8 pKa = 10.19KK9 pKa = 10.61RR10 pKa = 11.84FRR12 pKa = 11.84TTKK15 pKa = 10.0NGKK18 pKa = 9.75LIANKK23 pKa = 7.23TARR26 pKa = 11.84GHH28 pKa = 4.21MHH30 pKa = 6.63APKK33 pKa = 10.15NGKK36 pKa = 8.69ARR38 pKa = 11.84RR39 pKa = 11.84GKK41 pKa = 10.22RR42 pKa = 11.84KK43 pKa = 8.98PLVISGTWAVLMKK56 pKa = 10.94RR57 pKa = 11.84MMM59 pKa = 4.28
Molecular weight: 6.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1465947 |
32 |
2399 |
391.9 |
42.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.806 ± 0.055 | 0.883 ± 0.012 |
6.653 ± 0.036 | 6.665 ± 0.039 |
3.437 ± 0.024 | 9.256 ± 0.043 |
2.028 ± 0.019 | 3.368 ± 0.026 |
1.831 ± 0.027 | 10.738 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.593 ± 0.016 | 1.921 ± 0.024 |
5.502 ± 0.026 | 2.477 ± 0.022 |
8.393 ± 0.05 | 5.552 ± 0.027 |
5.326 ± 0.024 | 8.292 ± 0.038 |
1.489 ± 0.015 | 1.791 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
