
Pacific flying fox faeces associated gemycircularvirus-7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykolovirus; Gemykolovirus ptero2; Pteropus associated gemykolovirus 2
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
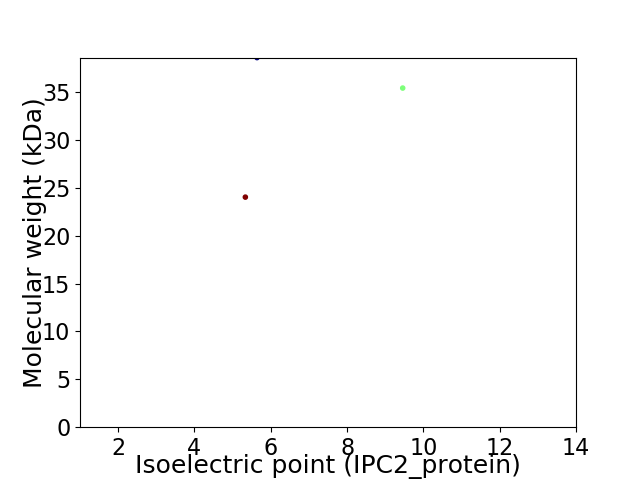
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTN4|A0A140CTN4_9VIRU RepA OS=Pacific flying fox faeces associated gemycircularvirus-7 OX=1795999 PE=4 SV=1
MM1 pKa = 6.83PAKK4 pKa = 10.46YY5 pKa = 10.31KK6 pKa = 10.68LDD8 pKa = 3.58NEE10 pKa = 4.36QFFMVTFVRR19 pKa = 11.84TPDD22 pKa = 3.35DD23 pKa = 3.64WPVCDD28 pKa = 6.22LITILDD34 pKa = 4.05RR35 pKa = 11.84LGCDD39 pKa = 3.17YY40 pKa = 11.01RR41 pKa = 11.84VGRR44 pKa = 11.84EE45 pKa = 3.6LHH47 pKa = 6.89ADD49 pKa = 4.2GKK51 pKa = 8.6PHH53 pKa = 5.13YY54 pKa = 10.11HH55 pKa = 6.52AMCVFDD61 pKa = 5.42EE62 pKa = 5.39PYY64 pKa = 10.89SDD66 pKa = 3.19TDD68 pKa = 3.13ARR70 pKa = 11.84KK71 pKa = 7.12TFQVAGRR78 pKa = 11.84SPNIRR83 pKa = 11.84VRR85 pKa = 11.84RR86 pKa = 11.84SRR88 pKa = 11.84PDD90 pKa = 3.07RR91 pKa = 11.84GWNYY95 pKa = 9.65VGKK98 pKa = 9.54HH99 pKa = 5.51AGTKK103 pKa = 9.22EE104 pKa = 3.67GHH106 pKa = 6.51EE107 pKa = 4.16IVAEE111 pKa = 4.1RR112 pKa = 11.84GEE114 pKa = 4.42CPMGDD119 pKa = 3.56EE120 pKa = 4.82EE121 pKa = 5.72DD122 pKa = 4.24RR123 pKa = 11.84PSNDD127 pKa = 2.35VWHH130 pKa = 6.84EE131 pKa = 3.99IILARR136 pKa = 11.84TRR138 pKa = 11.84EE139 pKa = 4.03EE140 pKa = 4.11FFEE143 pKa = 4.59KK144 pKa = 10.13CATMAPRR151 pKa = 11.84QLACCFGQLSSYY163 pKa = 11.23ADD165 pKa = 3.14WKK167 pKa = 8.81YY168 pKa = 11.32AKK170 pKa = 9.45PKK172 pKa = 9.86EE173 pKa = 4.47VYY175 pKa = 9.02EE176 pKa = 4.24PPVGEE181 pKa = 3.86FLEE184 pKa = 4.6IPEE187 pKa = 4.84EE188 pKa = 3.91LTSWVEE194 pKa = 4.26RR195 pKa = 11.84NLTNPNGRR203 pKa = 11.84YY204 pKa = 9.31VCII207 pKa = 4.32
MM1 pKa = 6.83PAKK4 pKa = 10.46YY5 pKa = 10.31KK6 pKa = 10.68LDD8 pKa = 3.58NEE10 pKa = 4.36QFFMVTFVRR19 pKa = 11.84TPDD22 pKa = 3.35DD23 pKa = 3.64WPVCDD28 pKa = 6.22LITILDD34 pKa = 4.05RR35 pKa = 11.84LGCDD39 pKa = 3.17YY40 pKa = 11.01RR41 pKa = 11.84VGRR44 pKa = 11.84EE45 pKa = 3.6LHH47 pKa = 6.89ADD49 pKa = 4.2GKK51 pKa = 8.6PHH53 pKa = 5.13YY54 pKa = 10.11HH55 pKa = 6.52AMCVFDD61 pKa = 5.42EE62 pKa = 5.39PYY64 pKa = 10.89SDD66 pKa = 3.19TDD68 pKa = 3.13ARR70 pKa = 11.84KK71 pKa = 7.12TFQVAGRR78 pKa = 11.84SPNIRR83 pKa = 11.84VRR85 pKa = 11.84RR86 pKa = 11.84SRR88 pKa = 11.84PDD90 pKa = 3.07RR91 pKa = 11.84GWNYY95 pKa = 9.65VGKK98 pKa = 9.54HH99 pKa = 5.51AGTKK103 pKa = 9.22EE104 pKa = 3.67GHH106 pKa = 6.51EE107 pKa = 4.16IVAEE111 pKa = 4.1RR112 pKa = 11.84GEE114 pKa = 4.42CPMGDD119 pKa = 3.56EE120 pKa = 4.82EE121 pKa = 5.72DD122 pKa = 4.24RR123 pKa = 11.84PSNDD127 pKa = 2.35VWHH130 pKa = 6.84EE131 pKa = 3.99IILARR136 pKa = 11.84TRR138 pKa = 11.84EE139 pKa = 4.03EE140 pKa = 4.11FFEE143 pKa = 4.59KK144 pKa = 10.13CATMAPRR151 pKa = 11.84QLACCFGQLSSYY163 pKa = 11.23ADD165 pKa = 3.14WKK167 pKa = 8.81YY168 pKa = 11.32AKK170 pKa = 9.45PKK172 pKa = 9.86EE173 pKa = 4.47VYY175 pKa = 9.02EE176 pKa = 4.24PPVGEE181 pKa = 3.86FLEE184 pKa = 4.6IPEE187 pKa = 4.84EE188 pKa = 3.91LTSWVEE194 pKa = 4.26RR195 pKa = 11.84NLTNPNGRR203 pKa = 11.84YY204 pKa = 9.31VCII207 pKa = 4.32
Molecular weight: 24.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTN3|A0A140CTN3_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated gemycircularvirus-7 OX=1795999 PE=4 SV=1
MM1 pKa = 7.91PYY3 pKa = 10.48GRR5 pKa = 11.84YY6 pKa = 9.09RR7 pKa = 11.84RR8 pKa = 11.84SSYY11 pKa = 10.18RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 10.12GRR16 pKa = 11.84SRR18 pKa = 11.84YY19 pKa = 9.71APRR22 pKa = 11.84SYY24 pKa = 10.88VRR26 pKa = 11.84RR27 pKa = 11.84PYY29 pKa = 10.54YY30 pKa = 9.73GVRR33 pKa = 11.84RR34 pKa = 11.84PFYY37 pKa = 9.15SRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 9.87RR43 pKa = 11.84PVSRR47 pKa = 11.84SILNVSSVKK56 pKa = 10.75KK57 pKa = 9.57KK58 pKa = 11.17DD59 pKa = 3.37NMLAVTNVLGTARR72 pKa = 11.84PSVQPFQDD80 pKa = 3.56GAASLVGSTAGRR92 pKa = 11.84GSPYY96 pKa = 10.45VFIWNATARR105 pKa = 11.84DD106 pKa = 4.0HH107 pKa = 7.23EE108 pKa = 5.05DD109 pKa = 3.84DD110 pKa = 3.52STKK113 pKa = 10.41YY114 pKa = 10.76DD115 pKa = 3.37EE116 pKa = 4.61AARR119 pKa = 11.84TSSAPYY125 pKa = 9.39MVGLKK130 pKa = 10.06EE131 pKa = 4.46RR132 pKa = 11.84ILVQTSDD139 pKa = 3.41ANPWQWRR146 pKa = 11.84RR147 pKa = 11.84ICFTAKK153 pKa = 10.22GLNSSFLTAGAGANEE168 pKa = 4.09NNPFYY173 pKa = 10.72EE174 pKa = 4.8DD175 pKa = 3.0STGFRR180 pKa = 11.84RR181 pKa = 11.84EE182 pKa = 3.73VAEE185 pKa = 4.62PGSAGLTNIRR195 pKa = 11.84NIIFDD200 pKa = 3.8GAYY203 pKa = 10.33GIDD206 pKa = 3.48WNDD209 pKa = 3.35PFSAKK214 pKa = 9.49TDD216 pKa = 3.34NQRR219 pKa = 11.84IKK221 pKa = 10.85VRR223 pKa = 11.84YY224 pKa = 9.2DD225 pKa = 2.88KK226 pKa = 11.24TRR228 pKa = 11.84MIRR231 pKa = 11.84STNNVGVIRR240 pKa = 11.84YY241 pKa = 9.1YY242 pKa = 11.65NMWHH246 pKa = 6.59PMKK249 pKa = 9.8KK250 pKa = 8.15TLMYY254 pKa = 10.94NDD256 pKa = 4.95DD257 pKa = 3.99EE258 pKa = 5.71NGASMDD264 pKa = 3.67SAYY267 pKa = 10.05VAQMGDD273 pKa = 3.36MGMGDD278 pKa = 4.13YY279 pKa = 10.6FVLDD283 pKa = 4.97FIRR286 pKa = 11.84CIDD289 pKa = 4.16APSGTPGLIFQPEE302 pKa = 3.88ATLYY306 pKa = 7.84WHH308 pKa = 6.69EE309 pKa = 4.18RR310 pKa = 3.47
MM1 pKa = 7.91PYY3 pKa = 10.48GRR5 pKa = 11.84YY6 pKa = 9.09RR7 pKa = 11.84RR8 pKa = 11.84SSYY11 pKa = 10.18RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 10.12GRR16 pKa = 11.84SRR18 pKa = 11.84YY19 pKa = 9.71APRR22 pKa = 11.84SYY24 pKa = 10.88VRR26 pKa = 11.84RR27 pKa = 11.84PYY29 pKa = 10.54YY30 pKa = 9.73GVRR33 pKa = 11.84RR34 pKa = 11.84PFYY37 pKa = 9.15SRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84YY42 pKa = 9.87RR43 pKa = 11.84PVSRR47 pKa = 11.84SILNVSSVKK56 pKa = 10.75KK57 pKa = 9.57KK58 pKa = 11.17DD59 pKa = 3.37NMLAVTNVLGTARR72 pKa = 11.84PSVQPFQDD80 pKa = 3.56GAASLVGSTAGRR92 pKa = 11.84GSPYY96 pKa = 10.45VFIWNATARR105 pKa = 11.84DD106 pKa = 4.0HH107 pKa = 7.23EE108 pKa = 5.05DD109 pKa = 3.84DD110 pKa = 3.52STKK113 pKa = 10.41YY114 pKa = 10.76DD115 pKa = 3.37EE116 pKa = 4.61AARR119 pKa = 11.84TSSAPYY125 pKa = 9.39MVGLKK130 pKa = 10.06EE131 pKa = 4.46RR132 pKa = 11.84ILVQTSDD139 pKa = 3.41ANPWQWRR146 pKa = 11.84RR147 pKa = 11.84ICFTAKK153 pKa = 10.22GLNSSFLTAGAGANEE168 pKa = 4.09NNPFYY173 pKa = 10.72EE174 pKa = 4.8DD175 pKa = 3.0STGFRR180 pKa = 11.84RR181 pKa = 11.84EE182 pKa = 3.73VAEE185 pKa = 4.62PGSAGLTNIRR195 pKa = 11.84NIIFDD200 pKa = 3.8GAYY203 pKa = 10.33GIDD206 pKa = 3.48WNDD209 pKa = 3.35PFSAKK214 pKa = 9.49TDD216 pKa = 3.34NQRR219 pKa = 11.84IKK221 pKa = 10.85VRR223 pKa = 11.84YY224 pKa = 9.2DD225 pKa = 2.88KK226 pKa = 11.24TRR228 pKa = 11.84MIRR231 pKa = 11.84STNNVGVIRR240 pKa = 11.84YY241 pKa = 9.1YY242 pKa = 11.65NMWHH246 pKa = 6.59PMKK249 pKa = 9.8KK250 pKa = 8.15TLMYY254 pKa = 10.94NDD256 pKa = 4.95DD257 pKa = 3.99EE258 pKa = 5.71NGASMDD264 pKa = 3.67SAYY267 pKa = 10.05VAQMGDD273 pKa = 3.36MGMGDD278 pKa = 4.13YY279 pKa = 10.6FVLDD283 pKa = 4.97FIRR286 pKa = 11.84CIDD289 pKa = 4.16APSGTPGLIFQPEE302 pKa = 3.88ATLYY306 pKa = 7.84WHH308 pKa = 6.69EE309 pKa = 4.18RR310 pKa = 3.47
Molecular weight: 35.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
848 |
207 |
331 |
282.7 |
32.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.722 ± 0.628 | 2.241 ± 0.837 |
7.075 ± 0.293 | 6.722 ± 1.736 |
4.245 ± 0.176 | 7.075 ± 0.373 |
2.358 ± 0.649 | 4.245 ± 0.165 |
5.071 ± 0.786 | 5.307 ± 0.548 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.066 ± 0.287 | 4.127 ± 0.784 |
6.25 ± 0.447 | 2.123 ± 0.082 |
8.844 ± 1.03 | 5.542 ± 1.385 |
5.307 ± 0.178 | 5.66 ± 0.46 |
2.83 ± 0.517 | 5.189 ± 0.891 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
