
Marispirochaeta aestuarii
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Marispirochaeta
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
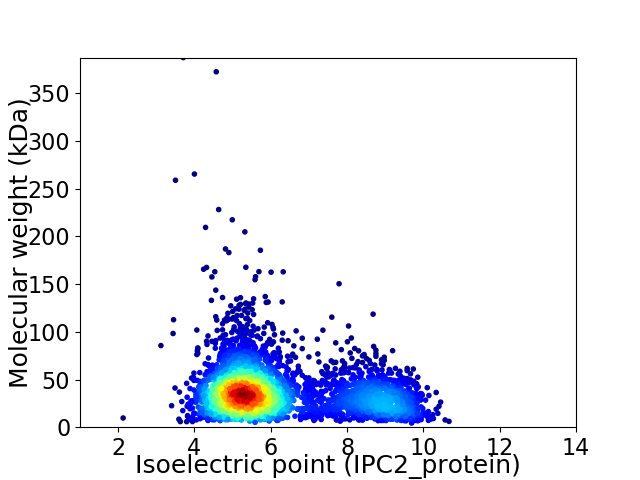
Virtual 2D-PAGE plot for 3763 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y1RYF4|A0A1Y1RYF4_9SPIO Chemotaxis protein OS=Marispirochaeta aestuarii OX=1963862 GN=B4O97_08160 PE=4 SV=1
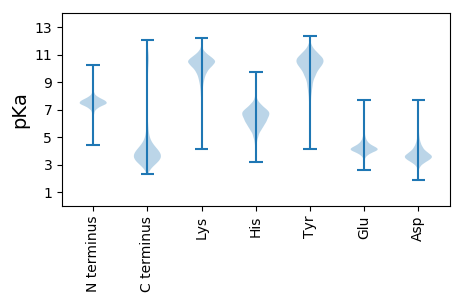
MM1 pKa = 7.29KK2 pKa = 10.2KK3 pKa = 9.84IVVVLVMLSLLVLPALAQTSFGLKK27 pKa = 9.94GALSDD32 pKa = 4.41YY33 pKa = 11.04SWTGDD38 pKa = 2.78GWGDD42 pKa = 3.38VQDD45 pKa = 4.5YY46 pKa = 10.14YY47 pKa = 11.47EE48 pKa = 4.24FMYY51 pKa = 10.53GVEE54 pKa = 4.01VDD56 pKa = 3.8NEE58 pKa = 4.04FSFGFSIGGFIEE70 pKa = 4.56HH71 pKa = 6.79KK72 pKa = 10.53VSSTLAIQPEE82 pKa = 4.56LLFTLASMQYY92 pKa = 11.11GDD94 pKa = 3.58GDD96 pKa = 3.46DD97 pKa = 4.44WIRR100 pKa = 11.84EE101 pKa = 4.03TWKK104 pKa = 9.55MLEE107 pKa = 3.71IPVYY111 pKa = 10.71LKK113 pKa = 11.19GLFPLDD119 pKa = 3.2QGSFYY124 pKa = 11.03IMGGPDD130 pKa = 3.94LFYY133 pKa = 11.26LLGDD137 pKa = 3.82IEE139 pKa = 5.6IDD141 pKa = 3.53TSNDD145 pKa = 3.1TSSSDD150 pKa = 4.42DD151 pKa = 4.66DD152 pKa = 4.41YY153 pKa = 11.99DD154 pKa = 4.17NNLLFGFAVSAGYY167 pKa = 9.86EE168 pKa = 4.12FQNGAFLGLKK178 pKa = 9.96FSRR181 pKa = 11.84VLTEE185 pKa = 4.58YY186 pKa = 11.42YY187 pKa = 10.72DD188 pKa = 3.85DD189 pKa = 3.72TDD191 pKa = 4.79LFIHH195 pKa = 7.18GIGIEE200 pKa = 4.13GGMKK204 pKa = 10.29LL205 pKa = 4.29
MM1 pKa = 7.29KK2 pKa = 10.2KK3 pKa = 9.84IVVVLVMLSLLVLPALAQTSFGLKK27 pKa = 9.94GALSDD32 pKa = 4.41YY33 pKa = 11.04SWTGDD38 pKa = 2.78GWGDD42 pKa = 3.38VQDD45 pKa = 4.5YY46 pKa = 10.14YY47 pKa = 11.47EE48 pKa = 4.24FMYY51 pKa = 10.53GVEE54 pKa = 4.01VDD56 pKa = 3.8NEE58 pKa = 4.04FSFGFSIGGFIEE70 pKa = 4.56HH71 pKa = 6.79KK72 pKa = 10.53VSSTLAIQPEE82 pKa = 4.56LLFTLASMQYY92 pKa = 11.11GDD94 pKa = 3.58GDD96 pKa = 3.46DD97 pKa = 4.44WIRR100 pKa = 11.84EE101 pKa = 4.03TWKK104 pKa = 9.55MLEE107 pKa = 3.71IPVYY111 pKa = 10.71LKK113 pKa = 11.19GLFPLDD119 pKa = 3.2QGSFYY124 pKa = 11.03IMGGPDD130 pKa = 3.94LFYY133 pKa = 11.26LLGDD137 pKa = 3.82IEE139 pKa = 5.6IDD141 pKa = 3.53TSNDD145 pKa = 3.1TSSSDD150 pKa = 4.42DD151 pKa = 4.66DD152 pKa = 4.41YY153 pKa = 11.99DD154 pKa = 4.17NNLLFGFAVSAGYY167 pKa = 9.86EE168 pKa = 4.12FQNGAFLGLKK178 pKa = 9.96FSRR181 pKa = 11.84VLTEE185 pKa = 4.58YY186 pKa = 11.42YY187 pKa = 10.72DD188 pKa = 3.85DD189 pKa = 3.72TDD191 pKa = 4.79LFIHH195 pKa = 7.18GIGIEE200 pKa = 4.13GGMKK204 pKa = 10.29LL205 pKa = 4.29
Molecular weight: 22.88 kDa
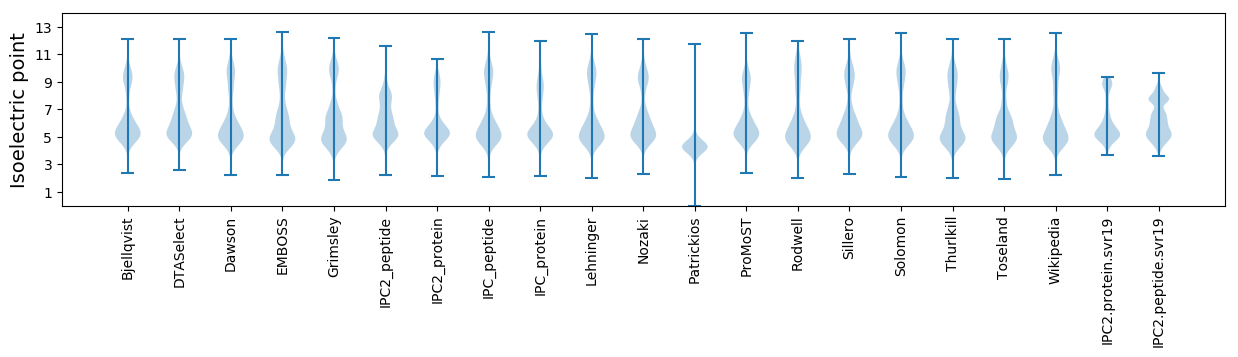
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y1S1U5|A0A1Y1S1U5_9SPIO Sugar ABC transporter substrate-binding protein OS=Marispirochaeta aestuarii OX=1963862 GN=B4O97_01735 PE=4 SV=1
MM1 pKa = 7.38NSCRR5 pKa = 11.84NISINLLRR13 pKa = 11.84WKK15 pKa = 9.4LCCGAALRR23 pKa = 11.84VAGINLRR30 pKa = 11.84YY31 pKa = 8.37LTAGTRR37 pKa = 11.84ILTLLSVSGCGSFKK51 pKa = 10.65QKK53 pKa = 10.17FRR55 pKa = 11.84FSRR58 pKa = 11.84RR59 pKa = 11.84VCTFHH64 pKa = 7.31GLTILIDD71 pKa = 3.62FVRR74 pKa = 11.84AARR77 pKa = 11.84TPSKK81 pKa = 10.01EE82 pKa = 3.56WKK84 pKa = 9.73LNLSAGEE91 pKa = 3.94DD92 pKa = 3.63TLNYY96 pKa = 9.83LHH98 pKa = 6.6SEE100 pKa = 3.86KK101 pKa = 10.53AIRR104 pKa = 11.84LQRR107 pKa = 11.84TIAKK111 pKa = 9.74HH112 pKa = 5.4FIEE115 pKa = 4.45RR116 pKa = 11.84QYY118 pKa = 11.32GIRR121 pKa = 11.84PEE123 pKa = 4.06PFEE126 pKa = 5.42LEE128 pKa = 4.23VLSRR132 pKa = 11.84CASVGAASGRR142 pKa = 11.84LIRR145 pKa = 11.84AAGCGSPTTLNRR157 pKa = 11.84EE158 pKa = 3.84DD159 pKa = 4.41LFRR162 pKa = 11.84CRR164 pKa = 11.84TVFPAFIRR172 pKa = 11.84LLNLDD177 pKa = 3.0MDD179 pKa = 4.05FRR181 pKa = 11.84EE182 pKa = 4.36RR183 pKa = 11.84NPVPEE188 pKa = 4.77PPLSRR193 pKa = 11.84FRR195 pKa = 11.84IKK197 pKa = 10.52LRR199 pKa = 11.84RR200 pKa = 11.84SLSLPRR206 pKa = 11.84RR207 pKa = 11.84TRR209 pKa = 11.84QPALPQLYY217 pKa = 9.13PRR219 pKa = 11.84EE220 pKa = 4.02LDD222 pKa = 3.18RR223 pKa = 11.84EE224 pKa = 3.9RR225 pKa = 11.84RR226 pKa = 11.84EE227 pKa = 3.95FEE229 pKa = 4.07RR230 pKa = 11.84TSLLARR236 pKa = 11.84YY237 pKa = 8.56CSSKK241 pKa = 10.83AILVRR246 pKa = 11.84TFTSEE251 pKa = 4.39LFRR254 pKa = 11.84LYY256 pKa = 10.56LDD258 pKa = 3.93AMQLIIEE265 pKa = 4.69NPPSKK270 pKa = 10.7AGLHH274 pKa = 5.95NIEE277 pKa = 4.58PLLNRR282 pKa = 11.84LEE284 pKa = 4.16RR285 pKa = 11.84MHH287 pKa = 6.56TKK289 pKa = 10.21SLNHH293 pKa = 5.87FRR295 pKa = 11.84EE296 pKa = 3.98EE297 pKa = 4.01NYY299 pKa = 10.93GYY301 pKa = 9.75MEE303 pKa = 4.25
MM1 pKa = 7.38NSCRR5 pKa = 11.84NISINLLRR13 pKa = 11.84WKK15 pKa = 9.4LCCGAALRR23 pKa = 11.84VAGINLRR30 pKa = 11.84YY31 pKa = 8.37LTAGTRR37 pKa = 11.84ILTLLSVSGCGSFKK51 pKa = 10.65QKK53 pKa = 10.17FRR55 pKa = 11.84FSRR58 pKa = 11.84RR59 pKa = 11.84VCTFHH64 pKa = 7.31GLTILIDD71 pKa = 3.62FVRR74 pKa = 11.84AARR77 pKa = 11.84TPSKK81 pKa = 10.01EE82 pKa = 3.56WKK84 pKa = 9.73LNLSAGEE91 pKa = 3.94DD92 pKa = 3.63TLNYY96 pKa = 9.83LHH98 pKa = 6.6SEE100 pKa = 3.86KK101 pKa = 10.53AIRR104 pKa = 11.84LQRR107 pKa = 11.84TIAKK111 pKa = 9.74HH112 pKa = 5.4FIEE115 pKa = 4.45RR116 pKa = 11.84QYY118 pKa = 11.32GIRR121 pKa = 11.84PEE123 pKa = 4.06PFEE126 pKa = 5.42LEE128 pKa = 4.23VLSRR132 pKa = 11.84CASVGAASGRR142 pKa = 11.84LIRR145 pKa = 11.84AAGCGSPTTLNRR157 pKa = 11.84EE158 pKa = 3.84DD159 pKa = 4.41LFRR162 pKa = 11.84CRR164 pKa = 11.84TVFPAFIRR172 pKa = 11.84LLNLDD177 pKa = 3.0MDD179 pKa = 4.05FRR181 pKa = 11.84EE182 pKa = 4.36RR183 pKa = 11.84NPVPEE188 pKa = 4.77PPLSRR193 pKa = 11.84FRR195 pKa = 11.84IKK197 pKa = 10.52LRR199 pKa = 11.84RR200 pKa = 11.84SLSLPRR206 pKa = 11.84RR207 pKa = 11.84TRR209 pKa = 11.84QPALPQLYY217 pKa = 9.13PRR219 pKa = 11.84EE220 pKa = 4.02LDD222 pKa = 3.18RR223 pKa = 11.84EE224 pKa = 3.9RR225 pKa = 11.84RR226 pKa = 11.84EE227 pKa = 3.95FEE229 pKa = 4.07RR230 pKa = 11.84TSLLARR236 pKa = 11.84YY237 pKa = 8.56CSSKK241 pKa = 10.83AILVRR246 pKa = 11.84TFTSEE251 pKa = 4.39LFRR254 pKa = 11.84LYY256 pKa = 10.56LDD258 pKa = 3.93AMQLIIEE265 pKa = 4.69NPPSKK270 pKa = 10.7AGLHH274 pKa = 5.95NIEE277 pKa = 4.58PLLNRR282 pKa = 11.84LEE284 pKa = 4.16RR285 pKa = 11.84MHH287 pKa = 6.56TKK289 pKa = 10.21SLNHH293 pKa = 5.87FRR295 pKa = 11.84EE296 pKa = 3.98EE297 pKa = 4.01NYY299 pKa = 10.93GYY301 pKa = 9.75MEE303 pKa = 4.25
Molecular weight: 35.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1298959 |
37 |
3619 |
345.2 |
38.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.054 ± 0.043 | 0.889 ± 0.014 |
5.467 ± 0.034 | 7.412 ± 0.045 |
4.473 ± 0.027 | 7.86 ± 0.034 |
1.773 ± 0.017 | 6.888 ± 0.042 |
4.537 ± 0.042 | 10.514 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.444 ± 0.02 | 3.433 ± 0.033 |
4.365 ± 0.028 | 2.912 ± 0.021 |
6.497 ± 0.039 | 6.808 ± 0.034 |
4.797 ± 0.028 | 6.603 ± 0.03 |
1.008 ± 0.013 | 3.268 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
