
Microviridae Fen7786_21
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
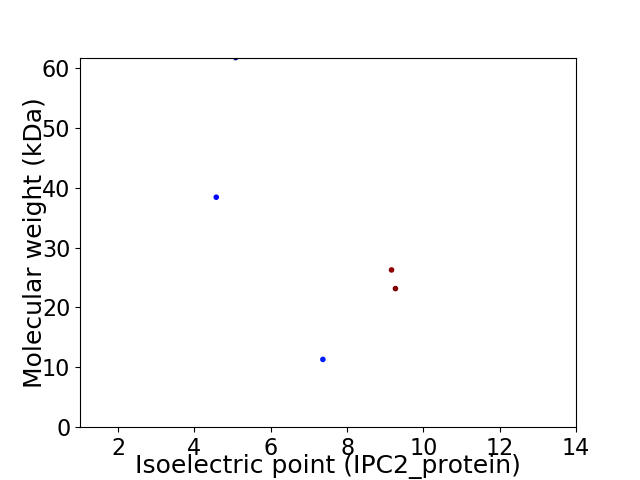
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UEG1|A0A0G2UEG1_9VIRU Uncharacterized protein OS=Microviridae Fen7786_21 OX=1655658 PE=4 SV=1
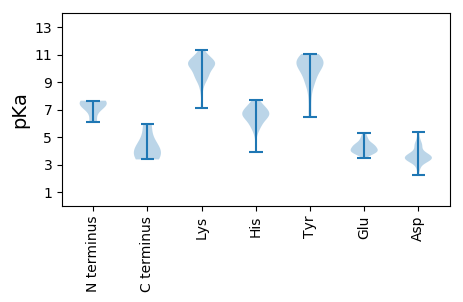
MM1 pKa = 7.64LIDD4 pKa = 3.8TSQFLSYY11 pKa = 10.33LKK13 pKa = 10.29HH14 pKa = 7.24SFMMYY19 pKa = 9.09LQVMASAGGLSAGQVLGAIGGALPVVGGIVSSLINNADD57 pKa = 3.4TTANNQANEE66 pKa = 4.0QFQLQQQQAQDD77 pKa = 3.5TYY79 pKa = 9.93NTQMWNNQNAYY90 pKa = 9.41NAPDD94 pKa = 3.35AQMSRR99 pKa = 11.84LEE101 pKa = 4.25DD102 pKa = 3.63AGLNPNLVYY111 pKa = 10.89GSGNVTGNVAEE122 pKa = 5.03AAPSMAKK129 pKa = 9.84VEE131 pKa = 4.4YY132 pKa = 10.64VSAPQRR138 pKa = 11.84APDD141 pKa = 3.83LASDD145 pKa = 4.37LTSGLAAYY153 pKa = 10.22NNLQLGSAQSDD164 pKa = 3.23NLAAQTHH171 pKa = 6.57LANSTSQLQSTEE183 pKa = 3.77AALNAASTDD192 pKa = 3.24KK193 pKa = 10.83TVAEE197 pKa = 4.56TSTIPAITAATVANMQSQTFKK218 pKa = 11.16NDD220 pKa = 2.2VDD222 pKa = 4.31SFVSSTLLPLQEE234 pKa = 4.64GQLSADD240 pKa = 3.4ATASLAAARR249 pKa = 11.84ANLTNAQQNVIKK261 pKa = 9.53TAVDD265 pKa = 3.54VKK267 pKa = 10.54TGIAQIAALAIQNAKK282 pKa = 9.98SQTEE286 pKa = 3.81IANIKK291 pKa = 9.66AQTALVLGNKK301 pKa = 9.25DD302 pKa = 3.75LQKK305 pKa = 10.18MDD307 pKa = 2.58ITMRR311 pKa = 11.84QQGEE315 pKa = 4.27MPNNSAAGNVIGAFATWLTGHH336 pKa = 6.02NQAGGYY342 pKa = 8.93NSNVTPNAIKK352 pKa = 10.69LSDD355 pKa = 3.64GTQINPDD362 pKa = 3.17GTTTKK367 pKa = 10.53QPP369 pKa = 3.37
MM1 pKa = 7.64LIDD4 pKa = 3.8TSQFLSYY11 pKa = 10.33LKK13 pKa = 10.29HH14 pKa = 7.24SFMMYY19 pKa = 9.09LQVMASAGGLSAGQVLGAIGGALPVVGGIVSSLINNADD57 pKa = 3.4TTANNQANEE66 pKa = 4.0QFQLQQQQAQDD77 pKa = 3.5TYY79 pKa = 9.93NTQMWNNQNAYY90 pKa = 9.41NAPDD94 pKa = 3.35AQMSRR99 pKa = 11.84LEE101 pKa = 4.25DD102 pKa = 3.63AGLNPNLVYY111 pKa = 10.89GSGNVTGNVAEE122 pKa = 5.03AAPSMAKK129 pKa = 9.84VEE131 pKa = 4.4YY132 pKa = 10.64VSAPQRR138 pKa = 11.84APDD141 pKa = 3.83LASDD145 pKa = 4.37LTSGLAAYY153 pKa = 10.22NNLQLGSAQSDD164 pKa = 3.23NLAAQTHH171 pKa = 6.57LANSTSQLQSTEE183 pKa = 3.77AALNAASTDD192 pKa = 3.24KK193 pKa = 10.83TVAEE197 pKa = 4.56TSTIPAITAATVANMQSQTFKK218 pKa = 11.16NDD220 pKa = 2.2VDD222 pKa = 4.31SFVSSTLLPLQEE234 pKa = 4.64GQLSADD240 pKa = 3.4ATASLAAARR249 pKa = 11.84ANLTNAQQNVIKK261 pKa = 9.53TAVDD265 pKa = 3.54VKK267 pKa = 10.54TGIAQIAALAIQNAKK282 pKa = 9.98SQTEE286 pKa = 3.81IANIKK291 pKa = 9.66AQTALVLGNKK301 pKa = 9.25DD302 pKa = 3.75LQKK305 pKa = 10.18MDD307 pKa = 2.58ITMRR311 pKa = 11.84QQGEE315 pKa = 4.27MPNNSAAGNVIGAFATWLTGHH336 pKa = 6.02NQAGGYY342 pKa = 8.93NSNVTPNAIKK352 pKa = 10.69LSDD355 pKa = 3.64GTQINPDD362 pKa = 3.17GTTTKK367 pKa = 10.53QPP369 pKa = 3.37
Molecular weight: 38.41 kDa
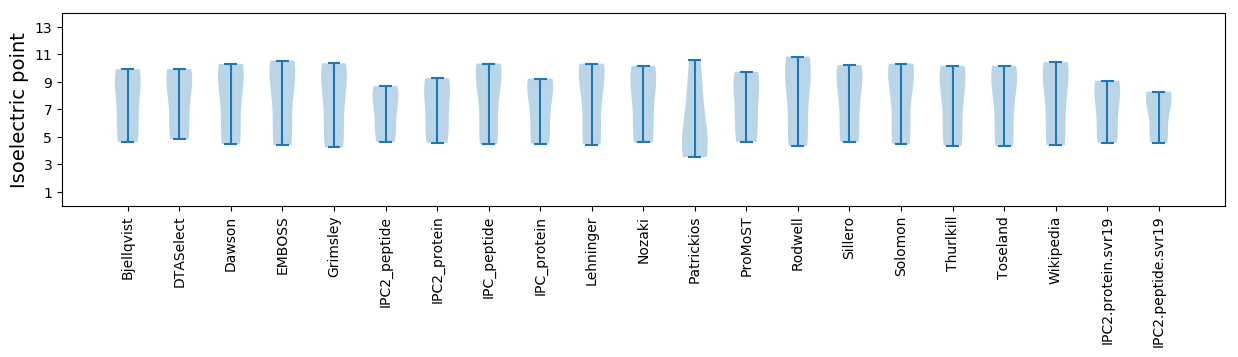
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UG51|A0A0G2UG51_9VIRU Uncharacterized protein OS=Microviridae Fen7786_21 OX=1655658 PE=4 SV=1
MM1 pKa = 7.26PTGSFPNRR9 pKa = 11.84IKK11 pKa = 9.93KK12 pKa = 7.14TCRR15 pKa = 11.84TCFRR19 pKa = 11.84LLLALLQRR27 pKa = 11.84LSSTNTFQFTVTLTISSALKK47 pKa = 8.94TKK49 pKa = 10.18PVTLLFVHH57 pKa = 6.59FNKK60 pKa = 10.38RR61 pKa = 11.84LNAAQINMKK70 pKa = 10.15KK71 pKa = 10.4ADD73 pKa = 3.38VHH75 pKa = 6.67SRR77 pKa = 11.84IVNHH81 pKa = 5.97LTYY84 pKa = 11.0SHH86 pKa = 7.45LEE88 pKa = 4.0TMEE91 pKa = 3.91LVLWKK96 pKa = 10.5RR97 pKa = 11.84SLTVPDD103 pKa = 3.5MVMPLSEE110 pKa = 4.32VIQRR114 pKa = 11.84YY115 pKa = 6.52TRR117 pKa = 11.84GQAVPTLPAVYY128 pKa = 10.44ADD130 pKa = 4.45IDD132 pKa = 3.98LSPYY136 pKa = 10.87DD137 pKa = 5.21DD138 pKa = 4.48LDD140 pKa = 3.45KK141 pKa = 11.34FQIIDD146 pKa = 3.61LQRR149 pKa = 11.84KK150 pKa = 8.24LEE152 pKa = 3.93HH153 pKa = 6.77RR154 pKa = 11.84ARR156 pKa = 11.84MLKK159 pKa = 9.42TDD161 pKa = 4.4IEE163 pKa = 4.28LTKK166 pKa = 11.16NKK168 pKa = 9.35IRR170 pKa = 11.84SQKK173 pKa = 10.21EE174 pKa = 3.45KK175 pKa = 10.06SDD177 pKa = 3.33YY178 pKa = 10.16DD179 pKa = 3.6ARR181 pKa = 11.84VAAAAKK187 pKa = 10.35ALSQTLNNDD196 pKa = 2.86NKK198 pKa = 9.35TQNSS202 pKa = 3.65
MM1 pKa = 7.26PTGSFPNRR9 pKa = 11.84IKK11 pKa = 9.93KK12 pKa = 7.14TCRR15 pKa = 11.84TCFRR19 pKa = 11.84LLLALLQRR27 pKa = 11.84LSSTNTFQFTVTLTISSALKK47 pKa = 8.94TKK49 pKa = 10.18PVTLLFVHH57 pKa = 6.59FNKK60 pKa = 10.38RR61 pKa = 11.84LNAAQINMKK70 pKa = 10.15KK71 pKa = 10.4ADD73 pKa = 3.38VHH75 pKa = 6.67SRR77 pKa = 11.84IVNHH81 pKa = 5.97LTYY84 pKa = 11.0SHH86 pKa = 7.45LEE88 pKa = 4.0TMEE91 pKa = 3.91LVLWKK96 pKa = 10.5RR97 pKa = 11.84SLTVPDD103 pKa = 3.5MVMPLSEE110 pKa = 4.32VIQRR114 pKa = 11.84YY115 pKa = 6.52TRR117 pKa = 11.84GQAVPTLPAVYY128 pKa = 10.44ADD130 pKa = 4.45IDD132 pKa = 3.98LSPYY136 pKa = 10.87DD137 pKa = 5.21DD138 pKa = 4.48LDD140 pKa = 3.45KK141 pKa = 11.34FQIIDD146 pKa = 3.61LQRR149 pKa = 11.84KK150 pKa = 8.24LEE152 pKa = 3.93HH153 pKa = 6.77RR154 pKa = 11.84ARR156 pKa = 11.84MLKK159 pKa = 9.42TDD161 pKa = 4.4IEE163 pKa = 4.28LTKK166 pKa = 11.16NKK168 pKa = 9.35IRR170 pKa = 11.84SQKK173 pKa = 10.21EE174 pKa = 3.45KK175 pKa = 10.06SDD177 pKa = 3.33YY178 pKa = 10.16DD179 pKa = 3.6ARR181 pKa = 11.84VAAAAKK187 pKa = 10.35ALSQTLNNDD196 pKa = 2.86NKK198 pKa = 9.35TQNSS202 pKa = 3.65
Molecular weight: 23.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1453 |
102 |
553 |
290.6 |
32.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.291 ± 2.308 | 1.101 ± 0.513 |
5.024 ± 0.392 | 3.923 ± 0.614 |
3.923 ± 0.965 | 5.988 ± 1.244 |
2.271 ± 0.498 | 5.437 ± 0.423 |
4.611 ± 1.341 | 9.566 ± 0.927 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.615 ± 0.158 | 5.85 ± 1.219 |
5.506 ± 0.986 | 5.919 ± 1.38 |
4.542 ± 1.314 | 7.158 ± 0.705 |
7.364 ± 0.655 | 5.299 ± 0.256 |
1.239 ± 0.35 | 3.372 ± 0.596 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
