
Alces alces faeces associated genomovirus MP43
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus lepam2
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
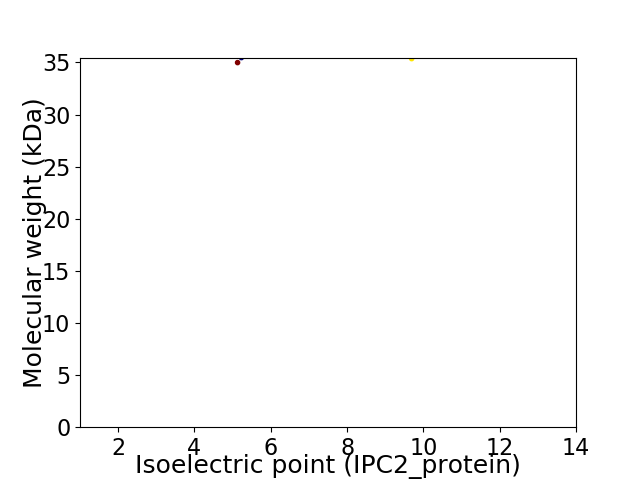
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
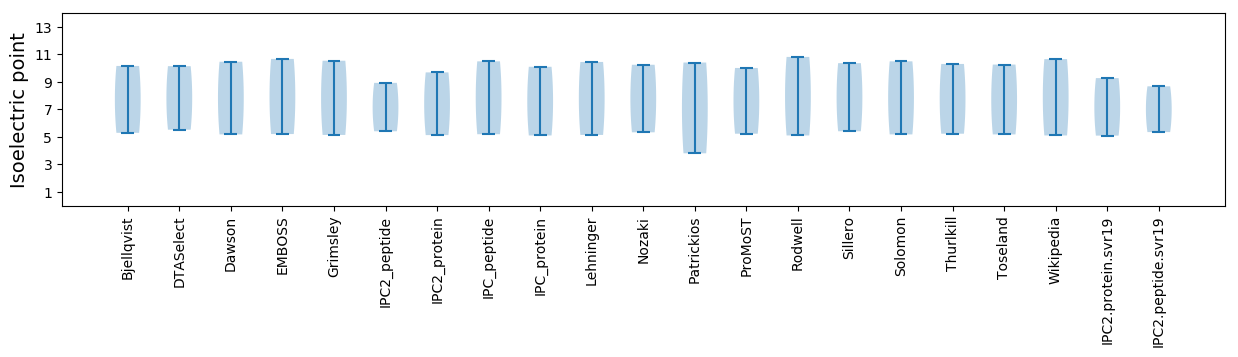
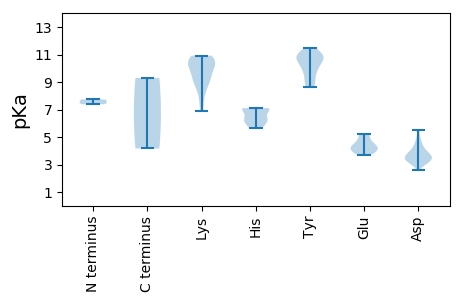
Protein with the lowest isoelectric point:
>tr|A0A2Z5CJA2|A0A2Z5CJA2_9VIRU Replication associated protein OS=Alces alces faeces associated genomovirus MP43 OX=2219114 PE=3 SV=1
MM1 pKa = 7.73PFVCNARR8 pKa = 11.84YY9 pKa = 8.93FLVTYY14 pKa = 8.73GHH16 pKa = 7.09VDD18 pKa = 3.09TLDD21 pKa = 3.61PFTIVDD27 pKa = 3.97FFGQLGAEE35 pKa = 4.26VIVGRR40 pKa = 11.84EE41 pKa = 4.0QYY43 pKa = 10.42HH44 pKa = 5.65ATLGTHH50 pKa = 5.68FHH52 pKa = 6.13VFADD56 pKa = 4.66FGRR59 pKa = 11.84KK60 pKa = 8.2FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84ADD67 pKa = 3.0IFDD70 pKa = 3.27VDD72 pKa = 4.66GYY74 pKa = 10.67HH75 pKa = 7.1PNISPSRR82 pKa = 11.84GTPEE86 pKa = 3.98AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSGVGPSGRR114 pKa = 11.84AAKK117 pKa = 8.64WHH119 pKa = 6.38SIIDD123 pKa = 3.51AEE125 pKa = 4.51TRR127 pKa = 11.84DD128 pKa = 3.84EE129 pKa = 5.15FYY131 pKa = 11.19SLCEE135 pKa = 3.81EE136 pKa = 4.97LDD138 pKa = 3.65PEE140 pKa = 4.71RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 9.8YY152 pKa = 10.1ADD154 pKa = 2.61WRR156 pKa = 11.84YY157 pKa = 9.15RR158 pKa = 11.84VEE160 pKa = 4.19PEE162 pKa = 4.6PYY164 pKa = 9.96ASPNGVFNLAEE175 pKa = 4.28YY176 pKa = 11.2GDD178 pKa = 4.16LSEE181 pKa = 4.74SKK183 pKa = 10.8SLILWGPSRR192 pKa = 11.84MGKK195 pKa = 6.86TVWARR200 pKa = 11.84SLGNHH205 pKa = 7.1LYY207 pKa = 10.71FGGIFSARR215 pKa = 11.84DD216 pKa = 3.28INRR219 pKa = 11.84DD220 pKa = 3.08GVEE223 pKa = 3.73YY224 pKa = 10.66AIFDD228 pKa = 5.13DD229 pKa = 3.53IAGGIKK235 pKa = 10.03FFPRR239 pKa = 11.84FKK241 pKa = 10.79DD242 pKa = 3.12WLGCQMEE249 pKa = 4.36FMVKK253 pKa = 9.32EE254 pKa = 4.5MYY256 pKa = 10.15RR257 pKa = 11.84DD258 pKa = 3.31PHH260 pKa = 6.37LFRR263 pKa = 11.84WGRR266 pKa = 11.84PAIWIANTDD275 pKa = 3.42PRR277 pKa = 11.84HH278 pKa = 6.13DD279 pKa = 3.82MSHH282 pKa = 6.98EE283 pKa = 4.0DD284 pKa = 3.94VTWLEE289 pKa = 4.04ANCIFVEE296 pKa = 3.75IDD298 pKa = 3.19SAIFHH303 pKa = 7.06ANTEE307 pKa = 4.17
MM1 pKa = 7.73PFVCNARR8 pKa = 11.84YY9 pKa = 8.93FLVTYY14 pKa = 8.73GHH16 pKa = 7.09VDD18 pKa = 3.09TLDD21 pKa = 3.61PFTIVDD27 pKa = 3.97FFGQLGAEE35 pKa = 4.26VIVGRR40 pKa = 11.84EE41 pKa = 4.0QYY43 pKa = 10.42HH44 pKa = 5.65ATLGTHH50 pKa = 5.68FHH52 pKa = 6.13VFADD56 pKa = 4.66FGRR59 pKa = 11.84KK60 pKa = 8.2FRR62 pKa = 11.84SRR64 pKa = 11.84RR65 pKa = 11.84ADD67 pKa = 3.0IFDD70 pKa = 3.27VDD72 pKa = 4.66GYY74 pKa = 10.67HH75 pKa = 7.1PNISPSRR82 pKa = 11.84GTPEE86 pKa = 3.98AGYY89 pKa = 10.62DD90 pKa = 3.63YY91 pKa = 10.68AIKK94 pKa = 10.92DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSGVGPSGRR114 pKa = 11.84AAKK117 pKa = 8.64WHH119 pKa = 6.38SIIDD123 pKa = 3.51AEE125 pKa = 4.51TRR127 pKa = 11.84DD128 pKa = 3.84EE129 pKa = 5.15FYY131 pKa = 11.19SLCEE135 pKa = 3.81EE136 pKa = 4.97LDD138 pKa = 3.65PEE140 pKa = 4.71RR141 pKa = 11.84LVCSFGQIQKK151 pKa = 9.8YY152 pKa = 10.1ADD154 pKa = 2.61WRR156 pKa = 11.84YY157 pKa = 9.15RR158 pKa = 11.84VEE160 pKa = 4.19PEE162 pKa = 4.6PYY164 pKa = 9.96ASPNGVFNLAEE175 pKa = 4.28YY176 pKa = 11.2GDD178 pKa = 4.16LSEE181 pKa = 4.74SKK183 pKa = 10.8SLILWGPSRR192 pKa = 11.84MGKK195 pKa = 6.86TVWARR200 pKa = 11.84SLGNHH205 pKa = 7.1LYY207 pKa = 10.71FGGIFSARR215 pKa = 11.84DD216 pKa = 3.28INRR219 pKa = 11.84DD220 pKa = 3.08GVEE223 pKa = 3.73YY224 pKa = 10.66AIFDD228 pKa = 5.13DD229 pKa = 3.53IAGGIKK235 pKa = 10.03FFPRR239 pKa = 11.84FKK241 pKa = 10.79DD242 pKa = 3.12WLGCQMEE249 pKa = 4.36FMVKK253 pKa = 9.32EE254 pKa = 4.5MYY256 pKa = 10.15RR257 pKa = 11.84DD258 pKa = 3.31PHH260 pKa = 6.37LFRR263 pKa = 11.84WGRR266 pKa = 11.84PAIWIANTDD275 pKa = 3.42PRR277 pKa = 11.84HH278 pKa = 6.13DD279 pKa = 3.82MSHH282 pKa = 6.98EE283 pKa = 4.0DD284 pKa = 3.94VTWLEE289 pKa = 4.04ANCIFVEE296 pKa = 3.75IDD298 pKa = 3.19SAIFHH303 pKa = 7.06ANTEE307 pKa = 4.17
Molecular weight: 34.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJA2|A0A2Z5CJA2_9VIRU Replication associated protein OS=Alces alces faeces associated genomovirus MP43 OX=2219114 PE=3 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84SRR5 pKa = 11.84YY6 pKa = 9.0YY7 pKa = 10.96GGGKK11 pKa = 7.6STGWANSKK19 pKa = 9.07RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.34YY23 pKa = 9.75RR24 pKa = 11.84SKK26 pKa = 10.61PRR28 pKa = 11.84RR29 pKa = 11.84SYY31 pKa = 10.85AKK33 pKa = 8.96KK34 pKa = 10.15RR35 pKa = 11.84SIFRR39 pKa = 11.84SKK41 pKa = 10.79RR42 pKa = 11.84MSTRR46 pKa = 11.84KK47 pKa = 9.41ILNITATKK55 pKa = 10.5KK56 pKa = 10.13RR57 pKa = 11.84DD58 pKa = 3.25TMMAWTNSSPSSQFGGATYY77 pKa = 10.09TNNAAVINGSAPDD90 pKa = 3.4STAQVFLWNATARR103 pKa = 11.84DD104 pKa = 3.62NSTSYY109 pKa = 11.02SAANPPTGSIFNTSTRR125 pKa = 11.84TSSNPYY131 pKa = 8.66MVGLAEE137 pKa = 5.19SIEE140 pKa = 4.26IQCNTGMPWQWRR152 pKa = 11.84RR153 pKa = 11.84ICFTMKK159 pKa = 10.4GPSLVPTSTASGAVFSTSAEE179 pKa = 4.14SSNGWLRR186 pKa = 11.84MMNQVGGNPGQNPMYY201 pKa = 10.65NLMVPLFRR209 pKa = 11.84GQVGADD215 pKa = 3.26WIDD218 pKa = 3.69PMTAATDD225 pKa = 3.59NSRR228 pKa = 11.84VTIKK232 pKa = 10.5YY233 pKa = 9.78DD234 pKa = 3.23KK235 pKa = 10.62TITLSSGNEE244 pKa = 3.75DD245 pKa = 3.51GFIRR249 pKa = 11.84TYY251 pKa = 10.94KK252 pKa = 9.53RR253 pKa = 11.84WHH255 pKa = 6.23PMKK258 pKa = 10.21STLVYY263 pKa = 10.9DD264 pKa = 4.67DD265 pKa = 5.5DD266 pKa = 4.23EE267 pKa = 5.18QGGSKK272 pKa = 8.76TVNIQSVSGKK282 pKa = 10.59AGMGDD287 pKa = 4.29YY288 pKa = 11.18YY289 pKa = 11.5VMDD292 pKa = 4.15MFRR295 pKa = 11.84ARR297 pKa = 11.84QGSAVADD304 pKa = 3.71QLTVRR309 pKa = 11.84PTATLYY315 pKa = 9.45WHH317 pKa = 7.01EE318 pKa = 4.29KK319 pKa = 9.3
MM1 pKa = 7.39ARR3 pKa = 11.84SRR5 pKa = 11.84YY6 pKa = 9.0YY7 pKa = 10.96GGGKK11 pKa = 7.6STGWANSKK19 pKa = 9.07RR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.34YY23 pKa = 9.75RR24 pKa = 11.84SKK26 pKa = 10.61PRR28 pKa = 11.84RR29 pKa = 11.84SYY31 pKa = 10.85AKK33 pKa = 8.96KK34 pKa = 10.15RR35 pKa = 11.84SIFRR39 pKa = 11.84SKK41 pKa = 10.79RR42 pKa = 11.84MSTRR46 pKa = 11.84KK47 pKa = 9.41ILNITATKK55 pKa = 10.5KK56 pKa = 10.13RR57 pKa = 11.84DD58 pKa = 3.25TMMAWTNSSPSSQFGGATYY77 pKa = 10.09TNNAAVINGSAPDD90 pKa = 3.4STAQVFLWNATARR103 pKa = 11.84DD104 pKa = 3.62NSTSYY109 pKa = 11.02SAANPPTGSIFNTSTRR125 pKa = 11.84TSSNPYY131 pKa = 8.66MVGLAEE137 pKa = 5.19SIEE140 pKa = 4.26IQCNTGMPWQWRR152 pKa = 11.84RR153 pKa = 11.84ICFTMKK159 pKa = 10.4GPSLVPTSTASGAVFSTSAEE179 pKa = 4.14SSNGWLRR186 pKa = 11.84MMNQVGGNPGQNPMYY201 pKa = 10.65NLMVPLFRR209 pKa = 11.84GQVGADD215 pKa = 3.26WIDD218 pKa = 3.69PMTAATDD225 pKa = 3.59NSRR228 pKa = 11.84VTIKK232 pKa = 10.5YY233 pKa = 9.78DD234 pKa = 3.23KK235 pKa = 10.62TITLSSGNEE244 pKa = 3.75DD245 pKa = 3.51GFIRR249 pKa = 11.84TYY251 pKa = 10.94KK252 pKa = 9.53RR253 pKa = 11.84WHH255 pKa = 6.23PMKK258 pKa = 10.21STLVYY263 pKa = 10.9DD264 pKa = 4.67DD265 pKa = 5.5DD266 pKa = 4.23EE267 pKa = 5.18QGGSKK272 pKa = 8.76TVNIQSVSGKK282 pKa = 10.59AGMGDD287 pKa = 4.29YY288 pKa = 11.18YY289 pKa = 11.5VMDD292 pKa = 4.15MFRR295 pKa = 11.84ARR297 pKa = 11.84QGSAVADD304 pKa = 3.71QLTVRR309 pKa = 11.84PTATLYY315 pKa = 9.45WHH317 pKa = 7.01EE318 pKa = 4.29KK319 pKa = 9.3
Molecular weight: 35.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
626 |
307 |
319 |
313.0 |
35.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.987 ± 0.114 | 1.118 ± 0.343 |
6.23 ± 1.284 | 3.994 ± 1.474 |
5.112 ± 1.598 | 8.626 ± 0.332 |
2.077 ± 1.011 | 5.112 ± 0.723 |
4.313 ± 0.927 | 4.473 ± 0.715 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.514 ± 1.047 | 4.633 ± 1.142 |
4.952 ± 0.174 | 2.556 ± 0.622 |
7.348 ± 0.122 | 8.626 ± 2.073 |
6.709 ± 2.099 | 5.431 ± 0.509 |
2.716 ± 0.074 | 4.473 ± 0.059 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
