
Rhodophyticola porphyridii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodophyticola
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
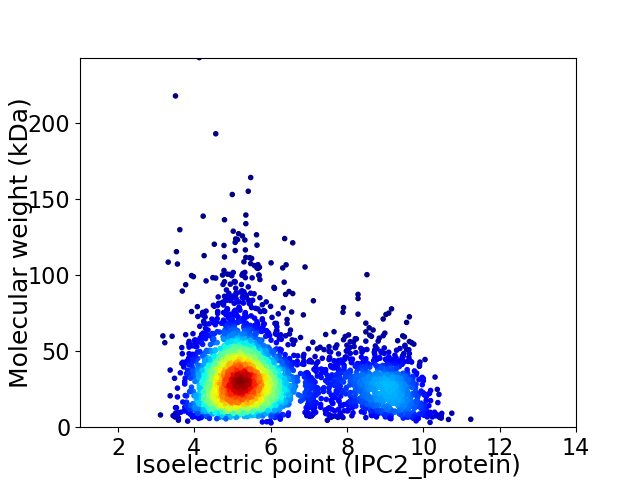
Virtual 2D-PAGE plot for 3660 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3L9Y173|A0A3L9Y173_9RHOB Integration host factor subunit alpha OS=Rhodophyticola porphyridii OX=1852017 GN=ihfA PE=3 SV=1
MM1 pKa = 7.48NFRR4 pKa = 11.84TGTFLTGAILAASALPAMAQNAEE27 pKa = 4.06LCGGVGAQGLWVGGSPEE44 pKa = 4.23ASDD47 pKa = 4.26ISTSDD52 pKa = 3.16TFIDD56 pKa = 3.59QTGLMVPPGGEE67 pKa = 4.18TVSLFSVSSAGEE79 pKa = 3.77YY80 pKa = 10.06RR81 pKa = 11.84IEE83 pKa = 4.33ANPNAGGDD91 pKa = 3.76TVIDD95 pKa = 3.53IRR97 pKa = 11.84DD98 pKa = 3.47EE99 pKa = 4.29AGTIILTDD107 pKa = 4.33DD108 pKa = 3.91DD109 pKa = 4.35SGGNFASRR117 pKa = 11.84GDD119 pKa = 3.44IALQPGTYY127 pKa = 9.57CVSTRR132 pKa = 11.84AFGGGAVVADD142 pKa = 3.86LRR144 pKa = 11.84VGRR147 pKa = 11.84AEE149 pKa = 4.62HH150 pKa = 6.39EE151 pKa = 4.12ALTPGWGGGIADD163 pKa = 4.46FAGIDD168 pKa = 3.52PCLPNTPATPLGAGAVDD185 pKa = 3.42AMLGEE190 pKa = 4.91GVTATNSIAGTPYY203 pKa = 10.54YY204 pKa = 10.1RR205 pKa = 11.84FSLSAPQALSIRR217 pKa = 11.84AEE219 pKa = 4.15NPSADD224 pKa = 3.86PYY226 pKa = 10.9IYY228 pKa = 9.86IYY230 pKa = 10.49DD231 pKa = 3.87GQGTLLAEE239 pKa = 4.31NDD241 pKa = 4.54DD242 pKa = 4.08YY243 pKa = 12.04DD244 pKa = 4.17GLNSRR249 pKa = 11.84VDD251 pKa = 3.99FTDD254 pKa = 3.86PLPAGNYY261 pKa = 8.36CIAMRR266 pKa = 11.84SLSDD270 pKa = 3.23DD271 pKa = 3.8TQPVTVSVRR280 pKa = 11.84GYY282 pKa = 10.77DD283 pKa = 3.29PAAAMQEE290 pKa = 4.02MFATGEE296 pKa = 3.9ASPPIGGSYY305 pKa = 10.16PITDD309 pKa = 4.1LGVLQTSLVMDD320 pKa = 3.31QVVGSDD326 pKa = 3.59AVWFSFSVPAGGLVLIDD343 pKa = 5.15AIEE346 pKa = 4.44ISDD349 pKa = 4.07SDD351 pKa = 3.79PVIRR355 pKa = 11.84LFDD358 pKa = 3.58GVGRR362 pKa = 11.84MIDD365 pKa = 3.75FNDD368 pKa = 4.25DD369 pKa = 3.5AGDD372 pKa = 3.86SLNSQLTVQVNPGTYY387 pKa = 8.92MLGVTQYY394 pKa = 9.73SEE396 pKa = 4.48SYY398 pKa = 10.09SGVIRR403 pKa = 11.84VALQRR408 pKa = 11.84YY409 pKa = 8.5VPAQQ413 pKa = 3.17
MM1 pKa = 7.48NFRR4 pKa = 11.84TGTFLTGAILAASALPAMAQNAEE27 pKa = 4.06LCGGVGAQGLWVGGSPEE44 pKa = 4.23ASDD47 pKa = 4.26ISTSDD52 pKa = 3.16TFIDD56 pKa = 3.59QTGLMVPPGGEE67 pKa = 4.18TVSLFSVSSAGEE79 pKa = 3.77YY80 pKa = 10.06RR81 pKa = 11.84IEE83 pKa = 4.33ANPNAGGDD91 pKa = 3.76TVIDD95 pKa = 3.53IRR97 pKa = 11.84DD98 pKa = 3.47EE99 pKa = 4.29AGTIILTDD107 pKa = 4.33DD108 pKa = 3.91DD109 pKa = 4.35SGGNFASRR117 pKa = 11.84GDD119 pKa = 3.44IALQPGTYY127 pKa = 9.57CVSTRR132 pKa = 11.84AFGGGAVVADD142 pKa = 3.86LRR144 pKa = 11.84VGRR147 pKa = 11.84AEE149 pKa = 4.62HH150 pKa = 6.39EE151 pKa = 4.12ALTPGWGGGIADD163 pKa = 4.46FAGIDD168 pKa = 3.52PCLPNTPATPLGAGAVDD185 pKa = 3.42AMLGEE190 pKa = 4.91GVTATNSIAGTPYY203 pKa = 10.54YY204 pKa = 10.1RR205 pKa = 11.84FSLSAPQALSIRR217 pKa = 11.84AEE219 pKa = 4.15NPSADD224 pKa = 3.86PYY226 pKa = 10.9IYY228 pKa = 9.86IYY230 pKa = 10.49DD231 pKa = 3.87GQGTLLAEE239 pKa = 4.31NDD241 pKa = 4.54DD242 pKa = 4.08YY243 pKa = 12.04DD244 pKa = 4.17GLNSRR249 pKa = 11.84VDD251 pKa = 3.99FTDD254 pKa = 3.86PLPAGNYY261 pKa = 8.36CIAMRR266 pKa = 11.84SLSDD270 pKa = 3.23DD271 pKa = 3.8TQPVTVSVRR280 pKa = 11.84GYY282 pKa = 10.77DD283 pKa = 3.29PAAAMQEE290 pKa = 4.02MFATGEE296 pKa = 3.9ASPPIGGSYY305 pKa = 10.16PITDD309 pKa = 4.1LGVLQTSLVMDD320 pKa = 3.31QVVGSDD326 pKa = 3.59AVWFSFSVPAGGLVLIDD343 pKa = 5.15AIEE346 pKa = 4.44ISDD349 pKa = 4.07SDD351 pKa = 3.79PVIRR355 pKa = 11.84LFDD358 pKa = 3.58GVGRR362 pKa = 11.84MIDD365 pKa = 3.75FNDD368 pKa = 4.25DD369 pKa = 3.5AGDD372 pKa = 3.86SLNSQLTVQVNPGTYY387 pKa = 8.92MLGVTQYY394 pKa = 9.73SEE396 pKa = 4.48SYY398 pKa = 10.09SGVIRR403 pKa = 11.84VALQRR408 pKa = 11.84YY409 pKa = 8.5VPAQQ413 pKa = 3.17
Molecular weight: 42.72 kDa
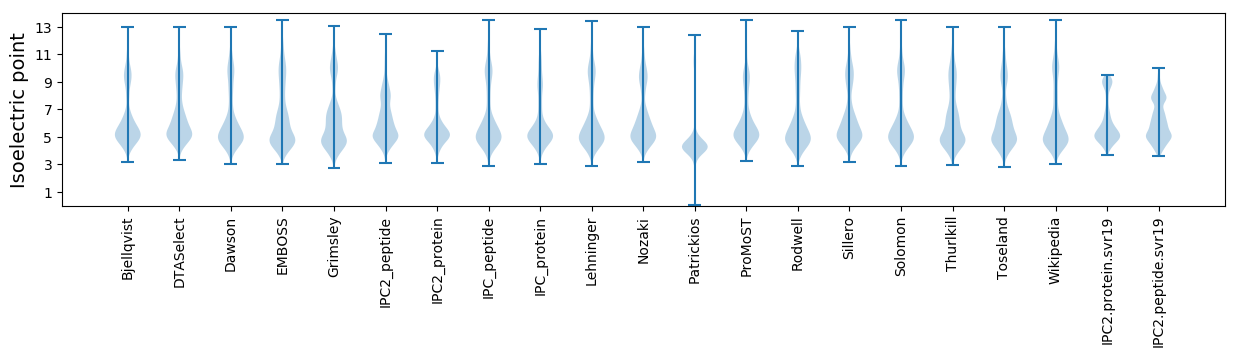
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3L9Y4M4|A0A3L9Y4M4_9RHOB GNAT family N-acetyltransferase OS=Rhodophyticola porphyridii OX=1852017 GN=D9R08_09805 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 9.18RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.07SLSAA45 pKa = 3.93
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 9.18RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.07SLSAA45 pKa = 3.93
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1138918 |
26 |
2388 |
311.2 |
33.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.589 ± 0.054 | 0.873 ± 0.012 |
6.136 ± 0.041 | 5.908 ± 0.039 |
3.722 ± 0.026 | 8.895 ± 0.046 |
2.1 ± 0.022 | 5.214 ± 0.03 |
2.523 ± 0.036 | 10.136 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.792 ± 0.019 | 2.426 ± 0.022 |
5.32 ± 0.031 | 3.014 ± 0.02 |
7.138 ± 0.047 | 4.966 ± 0.029 |
5.469 ± 0.026 | 7.176 ± 0.033 |
1.411 ± 0.019 | 2.192 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
