
candidate division MSBL1 archaeon SCGC-AAA259O05
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Euryarchaeota incertae sedis; candidate division MSBL1
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
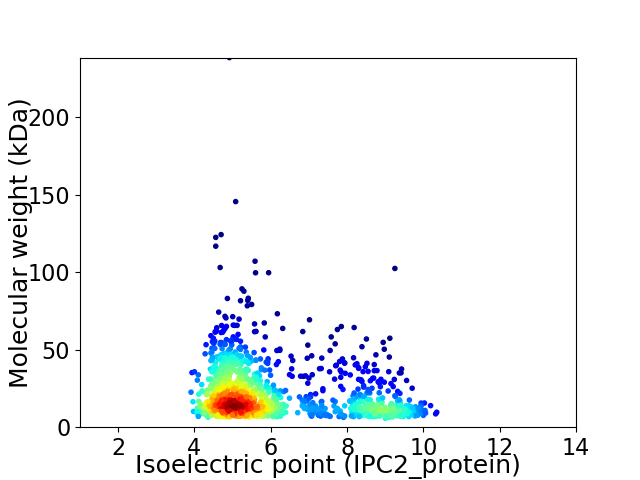
Virtual 2D-PAGE plot for 1138 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A133V3Q7|A0A133V3Q7_9EURY SPOB_a domain-containing protein OS=candidate division MSBL1 archaeon SCGC-AAA259O05 OX=1698271 GN=AKJ41_02830 PE=4 SV=1
MM1 pKa = 8.25KK2 pKa = 10.25YY3 pKa = 10.03RR4 pKa = 11.84QILILTFLIGLVAGLVPYY22 pKa = 10.94VMAVQSNNEE31 pKa = 3.9YY32 pKa = 10.86VGGFFKK38 pKa = 11.16NGGNKK43 pKa = 8.22YY44 pKa = 6.66TTDD47 pKa = 3.41KK48 pKa = 11.47AIMGNDD54 pKa = 3.19WDD56 pKa = 4.87NITEE60 pKa = 4.18GEE62 pKa = 4.25MKK64 pKa = 10.8YY65 pKa = 10.29IDD67 pKa = 3.95DD68 pKa = 3.97WVPAVLSVAGGNQQGDD84 pKa = 3.03WSGYY88 pKa = 8.05VYY90 pKa = 10.54QNPIHH95 pKa = 6.86LVRR98 pKa = 11.84DD99 pKa = 3.65DD100 pKa = 4.11SGAGGEE106 pKa = 4.32PLDD109 pKa = 6.4DD110 pKa = 3.64EE111 pKa = 5.96LFWNPQIWYY120 pKa = 9.77DD121 pKa = 5.3GNFLDD126 pKa = 5.42NKK128 pKa = 10.15GFLSTVIGTVNYY140 pKa = 6.06TTFYY144 pKa = 10.92VEE146 pKa = 4.62IRR148 pKa = 11.84GLSDD152 pKa = 4.07GYY154 pKa = 11.74ANYY157 pKa = 9.97KK158 pKa = 10.06AYY160 pKa = 10.07AYY162 pKa = 8.94DD163 pKa = 4.61TMDD166 pKa = 3.82QLINNNPYY174 pKa = 9.82VYY176 pKa = 9.46TWEE179 pKa = 3.87TDD181 pKa = 3.76YY182 pKa = 11.87YY183 pKa = 11.11SGDD186 pKa = 3.27SKK188 pKa = 11.73YY189 pKa = 10.99LVGTDD194 pKa = 3.2QGGGEE199 pKa = 4.23TLKK202 pKa = 10.63FFQFGVEE209 pKa = 4.19EE210 pKa = 4.21ASGTTWSSIGGEE222 pKa = 3.9WRR224 pKa = 11.84ISNYY228 pKa = 10.6NMGYY232 pKa = 10.14YY233 pKa = 10.33DD234 pKa = 4.63SSTNDD239 pKa = 2.46WRR241 pKa = 11.84YY242 pKa = 9.84LPGYY246 pKa = 9.12SVQHH250 pKa = 6.01EE251 pKa = 4.49DD252 pKa = 2.71SWITEE257 pKa = 4.45VEE259 pKa = 3.96GSPYY263 pKa = 10.54YY264 pKa = 10.13IGGQNYY270 pKa = 10.46DD271 pKa = 3.47GVHH274 pKa = 6.59IDD276 pKa = 3.73DD277 pKa = 4.13QGSDD281 pKa = 3.47FVMWQYY287 pKa = 11.62VEE289 pKa = 4.8GGSTVGSGVGLWSGEE304 pKa = 3.7GTYY307 pKa = 10.21IPDD310 pKa = 4.04PYY312 pKa = 10.89RR313 pKa = 5.42
MM1 pKa = 8.25KK2 pKa = 10.25YY3 pKa = 10.03RR4 pKa = 11.84QILILTFLIGLVAGLVPYY22 pKa = 10.94VMAVQSNNEE31 pKa = 3.9YY32 pKa = 10.86VGGFFKK38 pKa = 11.16NGGNKK43 pKa = 8.22YY44 pKa = 6.66TTDD47 pKa = 3.41KK48 pKa = 11.47AIMGNDD54 pKa = 3.19WDD56 pKa = 4.87NITEE60 pKa = 4.18GEE62 pKa = 4.25MKK64 pKa = 10.8YY65 pKa = 10.29IDD67 pKa = 3.95DD68 pKa = 3.97WVPAVLSVAGGNQQGDD84 pKa = 3.03WSGYY88 pKa = 8.05VYY90 pKa = 10.54QNPIHH95 pKa = 6.86LVRR98 pKa = 11.84DD99 pKa = 3.65DD100 pKa = 4.11SGAGGEE106 pKa = 4.32PLDD109 pKa = 6.4DD110 pKa = 3.64EE111 pKa = 5.96LFWNPQIWYY120 pKa = 9.77DD121 pKa = 5.3GNFLDD126 pKa = 5.42NKK128 pKa = 10.15GFLSTVIGTVNYY140 pKa = 6.06TTFYY144 pKa = 10.92VEE146 pKa = 4.62IRR148 pKa = 11.84GLSDD152 pKa = 4.07GYY154 pKa = 11.74ANYY157 pKa = 9.97KK158 pKa = 10.06AYY160 pKa = 10.07AYY162 pKa = 8.94DD163 pKa = 4.61TMDD166 pKa = 3.82QLINNNPYY174 pKa = 9.82VYY176 pKa = 9.46TWEE179 pKa = 3.87TDD181 pKa = 3.76YY182 pKa = 11.87YY183 pKa = 11.11SGDD186 pKa = 3.27SKK188 pKa = 11.73YY189 pKa = 10.99LVGTDD194 pKa = 3.2QGGGEE199 pKa = 4.23TLKK202 pKa = 10.63FFQFGVEE209 pKa = 4.19EE210 pKa = 4.21ASGTTWSSIGGEE222 pKa = 3.9WRR224 pKa = 11.84ISNYY228 pKa = 10.6NMGYY232 pKa = 10.14YY233 pKa = 10.33DD234 pKa = 4.63SSTNDD239 pKa = 2.46WRR241 pKa = 11.84YY242 pKa = 9.84LPGYY246 pKa = 9.12SVQHH250 pKa = 6.01EE251 pKa = 4.49DD252 pKa = 2.71SWITEE257 pKa = 4.45VEE259 pKa = 3.96GSPYY263 pKa = 10.54YY264 pKa = 10.13IGGQNYY270 pKa = 10.46DD271 pKa = 3.47GVHH274 pKa = 6.59IDD276 pKa = 3.73DD277 pKa = 4.13QGSDD281 pKa = 3.47FVMWQYY287 pKa = 11.62VEE289 pKa = 4.8GGSTVGSGVGLWSGEE304 pKa = 3.7GTYY307 pKa = 10.21IPDD310 pKa = 4.04PYY312 pKa = 10.89RR313 pKa = 5.42
Molecular weight: 35.21 kDa
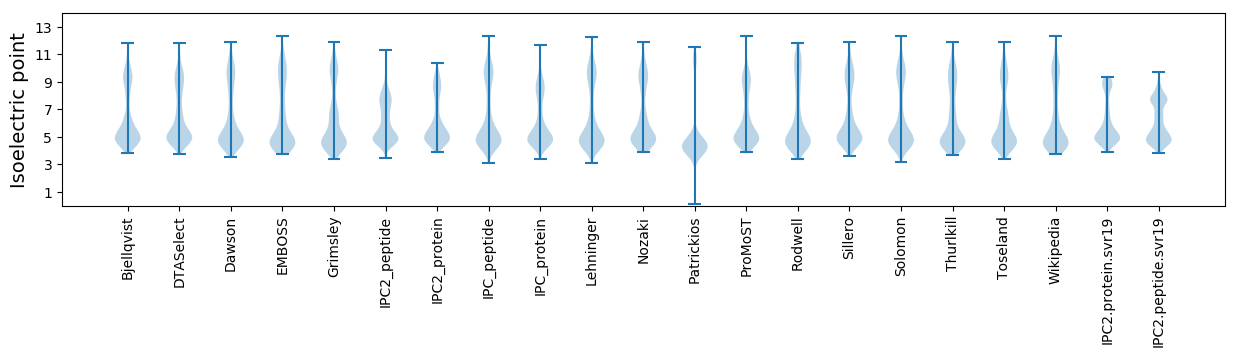
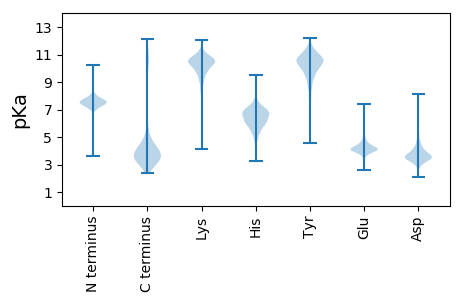
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133V4W1|A0A133V4W1_9EURY Probable tRNA sulfurtransferase OS=candidate division MSBL1 archaeon SCGC-AAA259O05 OX=1698271 GN=thiI PE=3 SV=1
MM1 pKa = 7.43GRR3 pKa = 11.84IRR5 pKa = 11.84SHH7 pKa = 6.21AGCRR11 pKa = 11.84FSRR14 pKa = 11.84PPVVEE19 pKa = 4.34RR20 pKa = 11.84FHH22 pKa = 8.33DD23 pKa = 4.26PDD25 pKa = 4.08SYY27 pKa = 11.57GHH29 pKa = 5.74GHH31 pKa = 6.91ASFFSRR37 pKa = 11.84LAAKK41 pKa = 7.61VTKK44 pKa = 10.35RR45 pKa = 11.84IQQRR49 pKa = 11.84RR50 pKa = 11.84SKK52 pKa = 10.38ISSLRR57 pKa = 11.84GKK59 pKa = 8.35ITRR62 pKa = 11.84SLGVQDD68 pKa = 4.49DD69 pKa = 4.31HH70 pKa = 7.37AALAPKK76 pKa = 9.83RR77 pKa = 11.84DD78 pKa = 3.42LWGEE82 pKa = 3.91DD83 pKa = 4.62DD84 pKa = 6.0IPLQTPLLYY93 pKa = 10.86GRR95 pKa = 11.84VGKK98 pKa = 10.27SINEE102 pKa = 3.52IGYY105 pKa = 7.59EE106 pKa = 3.95CRR108 pKa = 11.84KK109 pKa = 9.76RR110 pKa = 11.84VSKK113 pKa = 11.03
MM1 pKa = 7.43GRR3 pKa = 11.84IRR5 pKa = 11.84SHH7 pKa = 6.21AGCRR11 pKa = 11.84FSRR14 pKa = 11.84PPVVEE19 pKa = 4.34RR20 pKa = 11.84FHH22 pKa = 8.33DD23 pKa = 4.26PDD25 pKa = 4.08SYY27 pKa = 11.57GHH29 pKa = 5.74GHH31 pKa = 6.91ASFFSRR37 pKa = 11.84LAAKK41 pKa = 7.61VTKK44 pKa = 10.35RR45 pKa = 11.84IQQRR49 pKa = 11.84RR50 pKa = 11.84SKK52 pKa = 10.38ISSLRR57 pKa = 11.84GKK59 pKa = 8.35ITRR62 pKa = 11.84SLGVQDD68 pKa = 4.49DD69 pKa = 4.31HH70 pKa = 7.37AALAPKK76 pKa = 9.83RR77 pKa = 11.84DD78 pKa = 3.42LWGEE82 pKa = 3.91DD83 pKa = 4.62DD84 pKa = 6.0IPLQTPLLYY93 pKa = 10.86GRR95 pKa = 11.84VGKK98 pKa = 10.27SINEE102 pKa = 3.52IGYY105 pKa = 7.59EE106 pKa = 3.95CRR108 pKa = 11.84KK109 pKa = 9.76RR110 pKa = 11.84VSKK113 pKa = 11.03
Molecular weight: 12.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
238899 |
48 |
2078 |
209.9 |
23.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.031 ± 0.083 | 1.064 ± 0.03 |
5.775 ± 0.08 | 10.63 ± 0.114 |
3.966 ± 0.058 | 7.696 ± 0.077 |
1.796 ± 0.032 | 6.07 ± 0.069 |
7.141 ± 0.095 | 9.075 ± 0.108 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.177 ± 0.032 | 3.508 ± 0.049 |
4.342 ± 0.053 | 2.193 ± 0.029 |
6.292 ± 0.082 | 6.564 ± 0.07 |
4.504 ± 0.06 | 6.835 ± 0.061 |
1.329 ± 0.042 | 3.014 ± 0.053 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
