
Hubei tombus-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.88
Get precalculated fractions of proteins
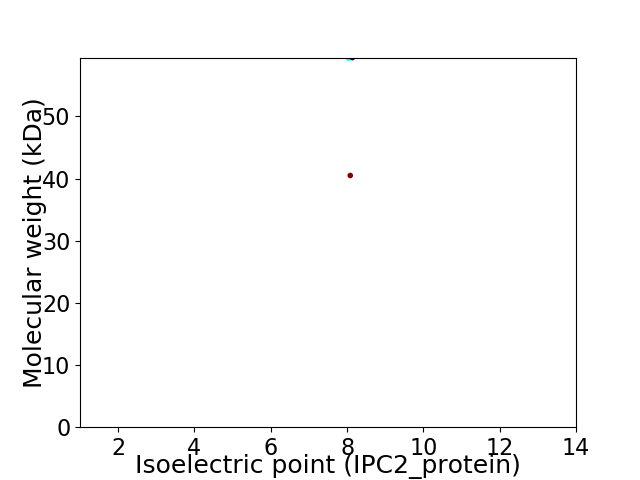
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGJ3|A0A1L3KGJ3_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 10 OX=1923256 PE=4 SV=1
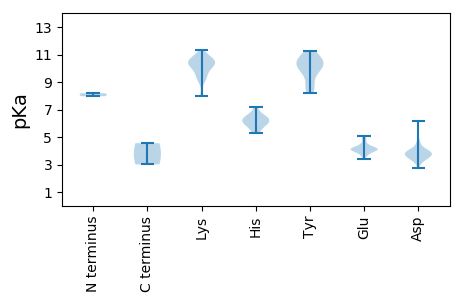
MM1 pKa = 8.19PDD3 pKa = 3.44LDD5 pKa = 3.72LGIRR9 pKa = 11.84VRR11 pKa = 11.84SFGSPIRR18 pKa = 11.84IRR20 pKa = 11.84RR21 pKa = 11.84STYY24 pKa = 8.58VSMVAGRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.69VFVHH37 pKa = 6.12NSSVHH42 pKa = 5.31NALLAAKK49 pKa = 9.75IRR51 pKa = 11.84VLTAEE56 pKa = 4.15VDD58 pKa = 3.77GKK60 pKa = 10.63LVPIEE65 pKa = 4.18EE66 pKa = 4.41TRR68 pKa = 11.84QPTGHH73 pKa = 5.62SHH75 pKa = 6.73ILDD78 pKa = 3.86GVLTTMQRR86 pKa = 11.84DD87 pKa = 3.93LPILTPVPLEE97 pKa = 4.02QFPGLYY103 pKa = 9.86KK104 pKa = 10.51AEE106 pKa = 4.05KK107 pKa = 9.26KK108 pKa = 10.37KK109 pKa = 10.37IYY111 pKa = 9.78EE112 pKa = 4.15RR113 pKa = 11.84AVSSLSVCPLTSDD126 pKa = 3.7DD127 pKa = 3.8SKK129 pKa = 11.14IRR131 pKa = 11.84AFIKK135 pKa = 10.12FEE137 pKa = 3.91KK138 pKa = 9.94MLAKK142 pKa = 9.43EE143 pKa = 4.12TGKK146 pKa = 9.95PKK148 pKa = 10.14APRR151 pKa = 11.84MISPPSPRR159 pKa = 11.84FLVRR163 pKa = 11.84TGCYY167 pKa = 9.46VKK169 pKa = 10.21PAEE172 pKa = 4.09HH173 pKa = 7.18AIYY176 pKa = 10.08EE177 pKa = 4.85AIDD180 pKa = 3.09HH181 pKa = 5.82MFGFKK186 pKa = 10.42VVTKK190 pKa = 10.24GMNFGEE196 pKa = 4.25IGRR199 pKa = 11.84LFKK202 pKa = 10.72SHH204 pKa = 6.39WDD206 pKa = 3.54ALDD209 pKa = 4.15DD210 pKa = 4.03PVAFDD215 pKa = 3.78VDD217 pKa = 4.0VEE219 pKa = 4.25KK220 pKa = 10.31MDD222 pKa = 4.1RR223 pKa = 11.84STSSEE228 pKa = 3.83MLAWTHH234 pKa = 6.39KK235 pKa = 10.4LIHH238 pKa = 6.0ACYY241 pKa = 10.13RR242 pKa = 11.84GDD244 pKa = 3.89DD245 pKa = 3.78LDD247 pKa = 6.15DD248 pKa = 3.47IRR250 pKa = 11.84EE251 pKa = 4.04MLKK254 pKa = 9.65QQLSVRR260 pKa = 11.84TTVKK264 pKa = 10.67CDD266 pKa = 3.5DD267 pKa = 3.94GNIKK271 pKa = 9.13YY272 pKa = 8.24TVDD275 pKa = 3.15GTLTSGQMNTSLVGVSMVSSIMYY298 pKa = 8.77TLFKK302 pKa = 10.69EE303 pKa = 4.36RR304 pKa = 11.84LDD306 pKa = 3.57VPYY309 pKa = 10.6RR310 pKa = 11.84FVDD313 pKa = 3.48AGDD316 pKa = 4.13DD317 pKa = 3.75CTVILDD323 pKa = 3.9KK324 pKa = 11.33RR325 pKa = 11.84NADD328 pKa = 3.56RR329 pKa = 11.84FLCEE333 pKa = 3.39VRR335 pKa = 11.84KK336 pKa = 10.17IFGEE340 pKa = 4.05VGFAITIGPPSVEE353 pKa = 3.81LEE355 pKa = 4.0QIEE358 pKa = 4.87FCQGHH363 pKa = 6.34PVLVGSSYY371 pKa = 11.06TMVRR375 pKa = 11.84NAKK378 pKa = 9.88DD379 pKa = 3.29AAIKK383 pKa = 10.46DD384 pKa = 3.93ATSLQPMDD392 pKa = 4.57SLRR395 pKa = 11.84EE396 pKa = 3.68MAVWMEE402 pKa = 3.98AVAKK406 pKa = 10.51CGIASHH412 pKa = 6.49GGVPIASSLYY422 pKa = 8.96RR423 pKa = 11.84CYY425 pKa = 11.14ARR427 pKa = 11.84NSTRR431 pKa = 11.84MQRR434 pKa = 11.84EE435 pKa = 3.61MKK437 pKa = 7.99MTQRR441 pKa = 11.84QLKK444 pKa = 9.04RR445 pKa = 11.84FKK447 pKa = 10.83LAVDD451 pKa = 3.38KK452 pKa = 10.95RR453 pKa = 11.84KK454 pKa = 10.29KK455 pKa = 10.62DD456 pKa = 3.59VVSWNQSNAEE466 pKa = 4.11PMCALDD472 pKa = 4.74VEE474 pKa = 4.66PSPYY478 pKa = 9.92TRR480 pKa = 11.84LSYY483 pKa = 10.99EE484 pKa = 3.79KK485 pKa = 10.82AFGLNSVFQLRR496 pKa = 11.84MEE498 pKa = 4.17QYY500 pKa = 11.27YY501 pKa = 8.56DD502 pKa = 3.32TLVINWSRR510 pKa = 11.84PVSQAQTTYY519 pKa = 10.17NPLWSTVV526 pKa = 3.01
MM1 pKa = 8.19PDD3 pKa = 3.44LDD5 pKa = 3.72LGIRR9 pKa = 11.84VRR11 pKa = 11.84SFGSPIRR18 pKa = 11.84IRR20 pKa = 11.84RR21 pKa = 11.84STYY24 pKa = 8.58VSMVAGRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.69VFVHH37 pKa = 6.12NSSVHH42 pKa = 5.31NALLAAKK49 pKa = 9.75IRR51 pKa = 11.84VLTAEE56 pKa = 4.15VDD58 pKa = 3.77GKK60 pKa = 10.63LVPIEE65 pKa = 4.18EE66 pKa = 4.41TRR68 pKa = 11.84QPTGHH73 pKa = 5.62SHH75 pKa = 6.73ILDD78 pKa = 3.86GVLTTMQRR86 pKa = 11.84DD87 pKa = 3.93LPILTPVPLEE97 pKa = 4.02QFPGLYY103 pKa = 9.86KK104 pKa = 10.51AEE106 pKa = 4.05KK107 pKa = 9.26KK108 pKa = 10.37KK109 pKa = 10.37IYY111 pKa = 9.78EE112 pKa = 4.15RR113 pKa = 11.84AVSSLSVCPLTSDD126 pKa = 3.7DD127 pKa = 3.8SKK129 pKa = 11.14IRR131 pKa = 11.84AFIKK135 pKa = 10.12FEE137 pKa = 3.91KK138 pKa = 9.94MLAKK142 pKa = 9.43EE143 pKa = 4.12TGKK146 pKa = 9.95PKK148 pKa = 10.14APRR151 pKa = 11.84MISPPSPRR159 pKa = 11.84FLVRR163 pKa = 11.84TGCYY167 pKa = 9.46VKK169 pKa = 10.21PAEE172 pKa = 4.09HH173 pKa = 7.18AIYY176 pKa = 10.08EE177 pKa = 4.85AIDD180 pKa = 3.09HH181 pKa = 5.82MFGFKK186 pKa = 10.42VVTKK190 pKa = 10.24GMNFGEE196 pKa = 4.25IGRR199 pKa = 11.84LFKK202 pKa = 10.72SHH204 pKa = 6.39WDD206 pKa = 3.54ALDD209 pKa = 4.15DD210 pKa = 4.03PVAFDD215 pKa = 3.78VDD217 pKa = 4.0VEE219 pKa = 4.25KK220 pKa = 10.31MDD222 pKa = 4.1RR223 pKa = 11.84STSSEE228 pKa = 3.83MLAWTHH234 pKa = 6.39KK235 pKa = 10.4LIHH238 pKa = 6.0ACYY241 pKa = 10.13RR242 pKa = 11.84GDD244 pKa = 3.89DD245 pKa = 3.78LDD247 pKa = 6.15DD248 pKa = 3.47IRR250 pKa = 11.84EE251 pKa = 4.04MLKK254 pKa = 9.65QQLSVRR260 pKa = 11.84TTVKK264 pKa = 10.67CDD266 pKa = 3.5DD267 pKa = 3.94GNIKK271 pKa = 9.13YY272 pKa = 8.24TVDD275 pKa = 3.15GTLTSGQMNTSLVGVSMVSSIMYY298 pKa = 8.77TLFKK302 pKa = 10.69EE303 pKa = 4.36RR304 pKa = 11.84LDD306 pKa = 3.57VPYY309 pKa = 10.6RR310 pKa = 11.84FVDD313 pKa = 3.48AGDD316 pKa = 4.13DD317 pKa = 3.75CTVILDD323 pKa = 3.9KK324 pKa = 11.33RR325 pKa = 11.84NADD328 pKa = 3.56RR329 pKa = 11.84FLCEE333 pKa = 3.39VRR335 pKa = 11.84KK336 pKa = 10.17IFGEE340 pKa = 4.05VGFAITIGPPSVEE353 pKa = 3.81LEE355 pKa = 4.0QIEE358 pKa = 4.87FCQGHH363 pKa = 6.34PVLVGSSYY371 pKa = 11.06TMVRR375 pKa = 11.84NAKK378 pKa = 9.88DD379 pKa = 3.29AAIKK383 pKa = 10.46DD384 pKa = 3.93ATSLQPMDD392 pKa = 4.57SLRR395 pKa = 11.84EE396 pKa = 3.68MAVWMEE402 pKa = 3.98AVAKK406 pKa = 10.51CGIASHH412 pKa = 6.49GGVPIASSLYY422 pKa = 8.96RR423 pKa = 11.84CYY425 pKa = 11.14ARR427 pKa = 11.84NSTRR431 pKa = 11.84MQRR434 pKa = 11.84EE435 pKa = 3.61MKK437 pKa = 7.99MTQRR441 pKa = 11.84QLKK444 pKa = 9.04RR445 pKa = 11.84FKK447 pKa = 10.83LAVDD451 pKa = 3.38KK452 pKa = 10.95RR453 pKa = 11.84KK454 pKa = 10.29KK455 pKa = 10.62DD456 pKa = 3.59VVSWNQSNAEE466 pKa = 4.11PMCALDD472 pKa = 4.74VEE474 pKa = 4.66PSPYY478 pKa = 9.92TRR480 pKa = 11.84LSYY483 pKa = 10.99EE484 pKa = 3.79KK485 pKa = 10.82AFGLNSVFQLRR496 pKa = 11.84MEE498 pKa = 4.17QYY500 pKa = 11.27YY501 pKa = 8.56DD502 pKa = 3.32TLVINWSRR510 pKa = 11.84PVSQAQTTYY519 pKa = 10.17NPLWSTVV526 pKa = 3.01
Molecular weight: 59.37 kDa
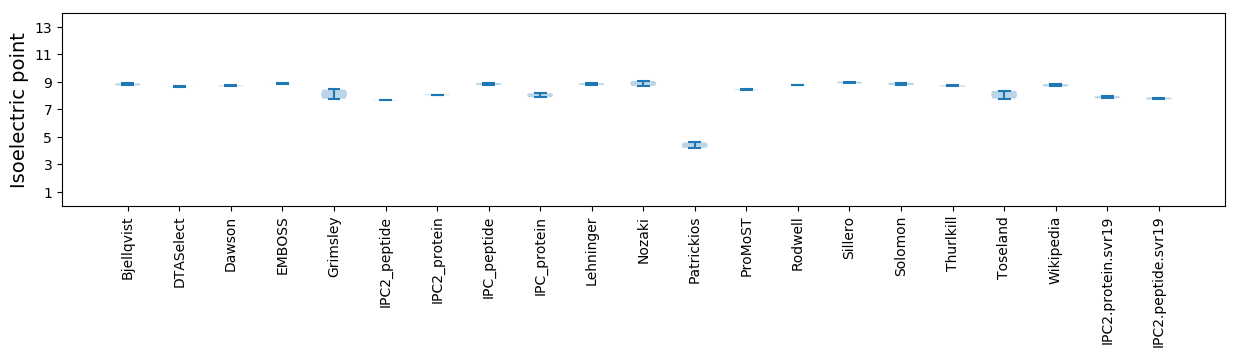
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGJ3|A0A1L3KGJ3_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 10 OX=1923256 PE=4 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84KK3 pKa = 9.38PSNLKK8 pKa = 9.97SANAWAKK15 pKa = 10.55SLSNPFGAPAAHH27 pKa = 6.7IPDD30 pKa = 4.54FEE32 pKa = 4.93TNSSGMITSTLYY44 pKa = 9.91TQVLPLGYY52 pKa = 10.45ASTATTHH59 pKa = 5.53NVGMIIQPHH68 pKa = 5.96PRR70 pKa = 11.84VHH72 pKa = 6.19FTQLSEE78 pKa = 4.49VNTGTANMTDD88 pKa = 3.66MNSGNTDD95 pKa = 2.71VGALLSVPNLAGFGDD110 pKa = 3.78QVRR113 pKa = 11.84YY114 pKa = 10.56RR115 pKa = 11.84MTSCGVRR122 pKa = 11.84VTYY125 pKa = 10.67AGTEE129 pKa = 4.07LNRR132 pKa = 11.84SGEE135 pKa = 4.25YY136 pKa = 9.8IAGFLQTDD144 pKa = 4.24YY145 pKa = 10.35PAQGTTAAATGIGPMSTLMPKK166 pKa = 8.01TTLGYY171 pKa = 8.2WTMAQIIGSLKK182 pKa = 10.25NPIEE186 pKa = 4.2ARR188 pKa = 11.84ICDD191 pKa = 3.94GTAEE195 pKa = 4.24FHH197 pKa = 6.33WKK199 pKa = 9.58PNGVPGYY206 pKa = 10.44GSGGANIAYY215 pKa = 9.43FPTTSGTAGAVTNPSGYY232 pKa = 10.71ASNPGEE238 pKa = 4.11GGVPYY243 pKa = 10.51GSEE246 pKa = 3.69NLVILIRR253 pKa = 11.84GDD255 pKa = 3.38TTAAAATTGNVYY267 pKa = 10.74DD268 pKa = 3.95VTVTSHH274 pKa = 6.1WEE276 pKa = 4.05VIPSVLNAVVYY287 pKa = 10.27DD288 pKa = 3.95VSPSLSDD295 pKa = 3.65PQALSAAMNVISRR308 pKa = 11.84RR309 pKa = 11.84PNPLFSTHH317 pKa = 5.95GVQEE321 pKa = 4.44TTFEE325 pKa = 4.11EE326 pKa = 3.97PVYY329 pKa = 10.46NVKK332 pKa = 10.36KK333 pKa = 10.82NKK335 pKa = 9.39VKK337 pKa = 10.72QIAQAAQEE345 pKa = 4.18SGLVQEE351 pKa = 5.07LGMRR355 pKa = 11.84ALRR358 pKa = 11.84VAAGIATRR366 pKa = 11.84KK367 pKa = 9.08LAGRR371 pKa = 11.84TRR373 pKa = 11.84APVPPGRR380 pKa = 11.84RR381 pKa = 11.84LEE383 pKa = 4.12FF384 pKa = 4.55
MM1 pKa = 7.97RR2 pKa = 11.84KK3 pKa = 9.38PSNLKK8 pKa = 9.97SANAWAKK15 pKa = 10.55SLSNPFGAPAAHH27 pKa = 6.7IPDD30 pKa = 4.54FEE32 pKa = 4.93TNSSGMITSTLYY44 pKa = 9.91TQVLPLGYY52 pKa = 10.45ASTATTHH59 pKa = 5.53NVGMIIQPHH68 pKa = 5.96PRR70 pKa = 11.84VHH72 pKa = 6.19FTQLSEE78 pKa = 4.49VNTGTANMTDD88 pKa = 3.66MNSGNTDD95 pKa = 2.71VGALLSVPNLAGFGDD110 pKa = 3.78QVRR113 pKa = 11.84YY114 pKa = 10.56RR115 pKa = 11.84MTSCGVRR122 pKa = 11.84VTYY125 pKa = 10.67AGTEE129 pKa = 4.07LNRR132 pKa = 11.84SGEE135 pKa = 4.25YY136 pKa = 9.8IAGFLQTDD144 pKa = 4.24YY145 pKa = 10.35PAQGTTAAATGIGPMSTLMPKK166 pKa = 8.01TTLGYY171 pKa = 8.2WTMAQIIGSLKK182 pKa = 10.25NPIEE186 pKa = 4.2ARR188 pKa = 11.84ICDD191 pKa = 3.94GTAEE195 pKa = 4.24FHH197 pKa = 6.33WKK199 pKa = 9.58PNGVPGYY206 pKa = 10.44GSGGANIAYY215 pKa = 9.43FPTTSGTAGAVTNPSGYY232 pKa = 10.71ASNPGEE238 pKa = 4.11GGVPYY243 pKa = 10.51GSEE246 pKa = 3.69NLVILIRR253 pKa = 11.84GDD255 pKa = 3.38TTAAAATTGNVYY267 pKa = 10.74DD268 pKa = 3.95VTVTSHH274 pKa = 6.1WEE276 pKa = 4.05VIPSVLNAVVYY287 pKa = 10.27DD288 pKa = 3.95VSPSLSDD295 pKa = 3.65PQALSAAMNVISRR308 pKa = 11.84RR309 pKa = 11.84PNPLFSTHH317 pKa = 5.95GVQEE321 pKa = 4.44TTFEE325 pKa = 4.11EE326 pKa = 3.97PVYY329 pKa = 10.46NVKK332 pKa = 10.36KK333 pKa = 10.82NKK335 pKa = 9.39VKK337 pKa = 10.72QIAQAAQEE345 pKa = 4.18SGLVQEE351 pKa = 5.07LGMRR355 pKa = 11.84ALRR358 pKa = 11.84VAAGIATRR366 pKa = 11.84KK367 pKa = 9.08LAGRR371 pKa = 11.84TRR373 pKa = 11.84APVPPGRR380 pKa = 11.84RR381 pKa = 11.84LEE383 pKa = 4.12FF384 pKa = 4.55
Molecular weight: 40.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
910 |
384 |
526 |
455.0 |
49.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.462 ± 1.53 | 1.319 ± 0.493 |
4.945 ± 1.447 | 4.615 ± 0.438 |
3.297 ± 0.428 | 7.363 ± 1.727 |
1.978 ± 0.096 | 4.835 ± 0.252 |
4.945 ± 1.286 | 7.363 ± 0.527 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.626 ± 0.471 | 3.956 ± 1.257 |
5.934 ± 0.678 | 3.297 ± 0.055 |
5.934 ± 0.931 | 7.692 ± 0.087 |
7.692 ± 1.523 | 8.242 ± 0.426 |
1.099 ± 0.035 | 3.407 ± 0.148 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
