
Streptomyces hainanensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
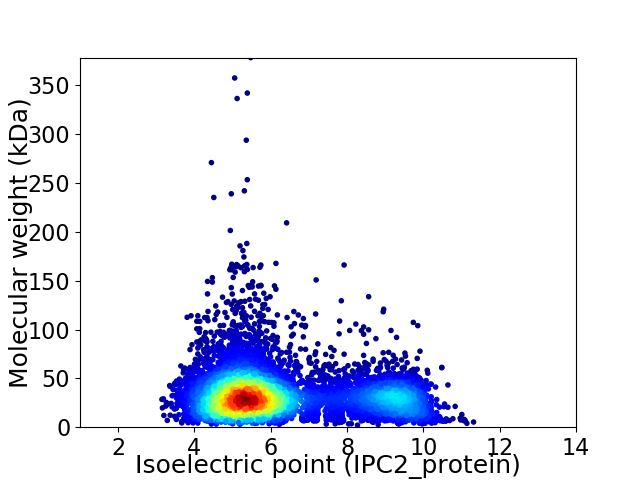
Virtual 2D-PAGE plot for 7007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R4T5P6|A0A4R4T5P6_9ACTN Ankyrin repeat domain-containing protein OS=Streptomyces hainanensis OX=402648 GN=E1283_21900 PE=4 SV=1
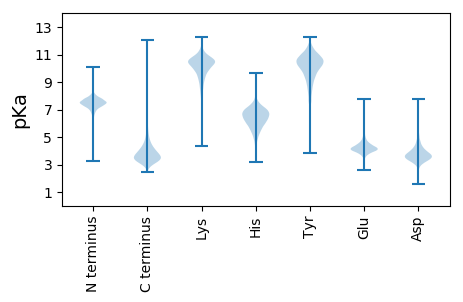
MM1 pKa = 7.01RR2 pKa = 11.84TARR5 pKa = 11.84PKK7 pKa = 9.52AAARR11 pKa = 11.84AFAAVIAIGLIAAGCASDD29 pKa = 5.81RR30 pKa = 11.84DD31 pKa = 4.18DD32 pKa = 6.19DD33 pKa = 5.27DD34 pKa = 6.26GSGDD38 pKa = 3.44GTGPFVFGAAGDD50 pKa = 3.96PAALDD55 pKa = 3.88PSLGSDD61 pKa = 3.48GEE63 pKa = 4.55TFRR66 pKa = 11.84VTRR69 pKa = 11.84QVFEE73 pKa = 4.08TLIEE77 pKa = 4.19HH78 pKa = 6.49EE79 pKa = 4.43PGGTEE84 pKa = 3.92MVPGLAEE91 pKa = 3.81EE92 pKa = 4.46WSSNEE97 pKa = 5.07AGTEE101 pKa = 3.75WSFTLRR107 pKa = 11.84DD108 pKa = 3.29GVTFHH113 pKa = 7.62DD114 pKa = 4.34GDD116 pKa = 3.96EE117 pKa = 4.3LTGEE121 pKa = 4.46VVCQNFEE128 pKa = 3.47RR129 pKa = 11.84WYY131 pKa = 8.91NWSGTYY137 pKa = 10.46QNTTLSYY144 pKa = 8.19YY145 pKa = 7.33WQSVFGGFAEE155 pKa = 4.36NEE157 pKa = 4.39SEE159 pKa = 4.24EE160 pKa = 4.62TPPPNYY166 pKa = 10.0VGCTAPDD173 pKa = 3.36EE174 pKa = 4.18LTAVIEE180 pKa = 4.26VAEE183 pKa = 4.49PSANYY188 pKa = 9.7PGGFSLQSFALQSPTAIEE206 pKa = 4.73AMAADD211 pKa = 4.61DD212 pKa = 4.22PSGEE216 pKa = 4.38GEE218 pKa = 4.55NITFPNYY225 pKa = 8.97SQEE228 pKa = 4.07AGVVSGTGPFQISEE242 pKa = 4.27WNRR245 pKa = 11.84SNGEE249 pKa = 3.61ITLEE253 pKa = 4.5RR254 pKa = 11.84FDD256 pKa = 6.09DD257 pKa = 3.94YY258 pKa = 11.2WGDD261 pKa = 3.35AAGVEE266 pKa = 4.19EE267 pKa = 5.01LVFRR271 pKa = 11.84TIPEE275 pKa = 3.58EE276 pKa = 3.83DD277 pKa = 3.56ARR279 pKa = 11.84RR280 pKa = 11.84QALQAGDD287 pKa = 3.29IDD289 pKa = 5.46GYY291 pKa = 11.37DD292 pKa = 3.52LVAPADD298 pKa = 3.92VEE300 pKa = 4.48TLVADD305 pKa = 4.86GFQVPVRR312 pKa = 11.84GVFNLLYY319 pKa = 10.82LGITQEE325 pKa = 4.15NNPALEE331 pKa = 4.01QLEE334 pKa = 4.25VRR336 pKa = 11.84QAIAHH341 pKa = 6.04ALDD344 pKa = 3.83RR345 pKa = 11.84EE346 pKa = 4.53NIVNTQLPEE355 pKa = 4.61GGVVATQFVPDD366 pKa = 4.72TIDD369 pKa = 3.82GYY371 pKa = 11.53SDD373 pKa = 4.77DD374 pKa = 3.92VTTYY378 pKa = 10.6EE379 pKa = 4.9HH380 pKa = 7.43DD381 pKa = 4.06PEE383 pKa = 5.1LAQQLLADD391 pKa = 5.09AGAEE395 pKa = 3.84DD396 pKa = 4.25LTIDD400 pKa = 3.97FCYY403 pKa = 8.49PTEE406 pKa = 3.95VTRR409 pKa = 11.84PYY411 pKa = 9.77MPAPADD417 pKa = 3.09IYY419 pKa = 11.11EE420 pKa = 4.37LLKK423 pKa = 10.8ADD425 pKa = 4.72LEE427 pKa = 4.58AVGITVNPVALKK439 pKa = 9.74WNPDD443 pKa = 3.4YY444 pKa = 11.65TEE446 pKa = 3.91TTRR449 pKa = 11.84DD450 pKa = 3.65GGCGVYY456 pKa = 10.81LLGWTGDD463 pKa = 3.53FNDD466 pKa = 4.77AYY468 pKa = 10.95NFLGTWFADD477 pKa = 4.11YY478 pKa = 10.68SRR480 pKa = 11.84EE481 pKa = 3.93WGFEE485 pKa = 3.89NQEE488 pKa = 4.16LFDD491 pKa = 3.99MMSAASTEE499 pKa = 4.01PDD501 pKa = 3.08VATRR505 pKa = 11.84VSLYY509 pKa = 10.45QGLNEE514 pKa = 4.72YY515 pKa = 10.56IMDD518 pKa = 4.32YY519 pKa = 10.93LPGVPISSSPPSIAFSPDD537 pKa = 2.95VNPPTVSPLTQEE549 pKa = 3.9NFAEE553 pKa = 4.69VTFKK557 pKa = 11.22
MM1 pKa = 7.01RR2 pKa = 11.84TARR5 pKa = 11.84PKK7 pKa = 9.52AAARR11 pKa = 11.84AFAAVIAIGLIAAGCASDD29 pKa = 5.81RR30 pKa = 11.84DD31 pKa = 4.18DD32 pKa = 6.19DD33 pKa = 5.27DD34 pKa = 6.26GSGDD38 pKa = 3.44GTGPFVFGAAGDD50 pKa = 3.96PAALDD55 pKa = 3.88PSLGSDD61 pKa = 3.48GEE63 pKa = 4.55TFRR66 pKa = 11.84VTRR69 pKa = 11.84QVFEE73 pKa = 4.08TLIEE77 pKa = 4.19HH78 pKa = 6.49EE79 pKa = 4.43PGGTEE84 pKa = 3.92MVPGLAEE91 pKa = 3.81EE92 pKa = 4.46WSSNEE97 pKa = 5.07AGTEE101 pKa = 3.75WSFTLRR107 pKa = 11.84DD108 pKa = 3.29GVTFHH113 pKa = 7.62DD114 pKa = 4.34GDD116 pKa = 3.96EE117 pKa = 4.3LTGEE121 pKa = 4.46VVCQNFEE128 pKa = 3.47RR129 pKa = 11.84WYY131 pKa = 8.91NWSGTYY137 pKa = 10.46QNTTLSYY144 pKa = 8.19YY145 pKa = 7.33WQSVFGGFAEE155 pKa = 4.36NEE157 pKa = 4.39SEE159 pKa = 4.24EE160 pKa = 4.62TPPPNYY166 pKa = 10.0VGCTAPDD173 pKa = 3.36EE174 pKa = 4.18LTAVIEE180 pKa = 4.26VAEE183 pKa = 4.49PSANYY188 pKa = 9.7PGGFSLQSFALQSPTAIEE206 pKa = 4.73AMAADD211 pKa = 4.61DD212 pKa = 4.22PSGEE216 pKa = 4.38GEE218 pKa = 4.55NITFPNYY225 pKa = 8.97SQEE228 pKa = 4.07AGVVSGTGPFQISEE242 pKa = 4.27WNRR245 pKa = 11.84SNGEE249 pKa = 3.61ITLEE253 pKa = 4.5RR254 pKa = 11.84FDD256 pKa = 6.09DD257 pKa = 3.94YY258 pKa = 11.2WGDD261 pKa = 3.35AAGVEE266 pKa = 4.19EE267 pKa = 5.01LVFRR271 pKa = 11.84TIPEE275 pKa = 3.58EE276 pKa = 3.83DD277 pKa = 3.56ARR279 pKa = 11.84RR280 pKa = 11.84QALQAGDD287 pKa = 3.29IDD289 pKa = 5.46GYY291 pKa = 11.37DD292 pKa = 3.52LVAPADD298 pKa = 3.92VEE300 pKa = 4.48TLVADD305 pKa = 4.86GFQVPVRR312 pKa = 11.84GVFNLLYY319 pKa = 10.82LGITQEE325 pKa = 4.15NNPALEE331 pKa = 4.01QLEE334 pKa = 4.25VRR336 pKa = 11.84QAIAHH341 pKa = 6.04ALDD344 pKa = 3.83RR345 pKa = 11.84EE346 pKa = 4.53NIVNTQLPEE355 pKa = 4.61GGVVATQFVPDD366 pKa = 4.72TIDD369 pKa = 3.82GYY371 pKa = 11.53SDD373 pKa = 4.77DD374 pKa = 3.92VTTYY378 pKa = 10.6EE379 pKa = 4.9HH380 pKa = 7.43DD381 pKa = 4.06PEE383 pKa = 5.1LAQQLLADD391 pKa = 5.09AGAEE395 pKa = 3.84DD396 pKa = 4.25LTIDD400 pKa = 3.97FCYY403 pKa = 8.49PTEE406 pKa = 3.95VTRR409 pKa = 11.84PYY411 pKa = 9.77MPAPADD417 pKa = 3.09IYY419 pKa = 11.11EE420 pKa = 4.37LLKK423 pKa = 10.8ADD425 pKa = 4.72LEE427 pKa = 4.58AVGITVNPVALKK439 pKa = 9.74WNPDD443 pKa = 3.4YY444 pKa = 11.65TEE446 pKa = 3.91TTRR449 pKa = 11.84DD450 pKa = 3.65GGCGVYY456 pKa = 10.81LLGWTGDD463 pKa = 3.53FNDD466 pKa = 4.77AYY468 pKa = 10.95NFLGTWFADD477 pKa = 4.11YY478 pKa = 10.68SRR480 pKa = 11.84EE481 pKa = 3.93WGFEE485 pKa = 3.89NQEE488 pKa = 4.16LFDD491 pKa = 3.99MMSAASTEE499 pKa = 4.01PDD501 pKa = 3.08VATRR505 pKa = 11.84VSLYY509 pKa = 10.45QGLNEE514 pKa = 4.72YY515 pKa = 10.56IMDD518 pKa = 4.32YY519 pKa = 10.93LPGVPISSSPPSIAFSPDD537 pKa = 2.95VNPPTVSPLTQEE549 pKa = 3.9NFAEE553 pKa = 4.69VTFKK557 pKa = 11.22
Molecular weight: 60.54 kDa
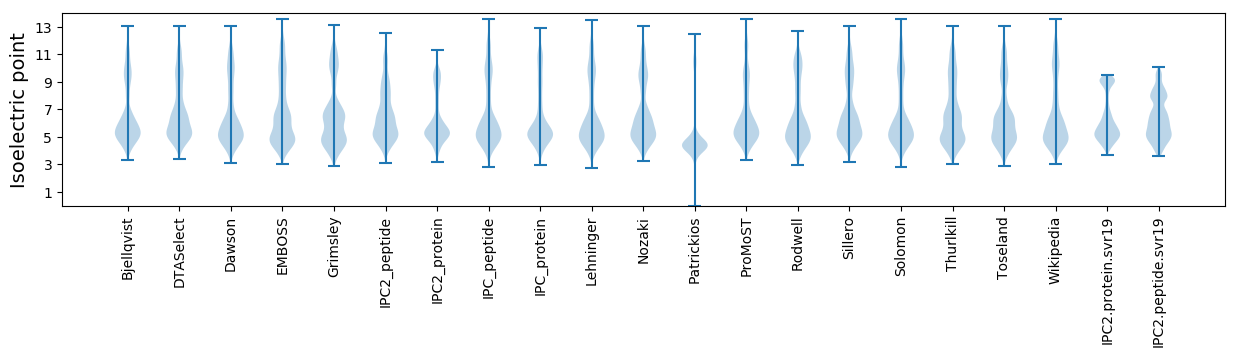
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R4T9F4|A0A4R4T9F4_9ACTN GAF domain-containing protein OS=Streptomyces hainanensis OX=402648 GN=E1283_22705 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.43GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.43GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2299493 |
18 |
3673 |
328.2 |
35.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.076 ± 0.05 | 0.725 ± 0.007 |
6.117 ± 0.025 | 5.862 ± 0.029 |
2.714 ± 0.017 | 9.642 ± 0.027 |
2.296 ± 0.015 | 2.981 ± 0.021 |
1.217 ± 0.016 | 10.73 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.608 ± 0.013 | 1.704 ± 0.018 |
6.272 ± 0.026 | 2.506 ± 0.018 |
8.736 ± 0.029 | 4.738 ± 0.021 |
6.043 ± 0.025 | 8.505 ± 0.026 |
1.555 ± 0.013 | 1.974 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
