
Microviridae Fen7895_21
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
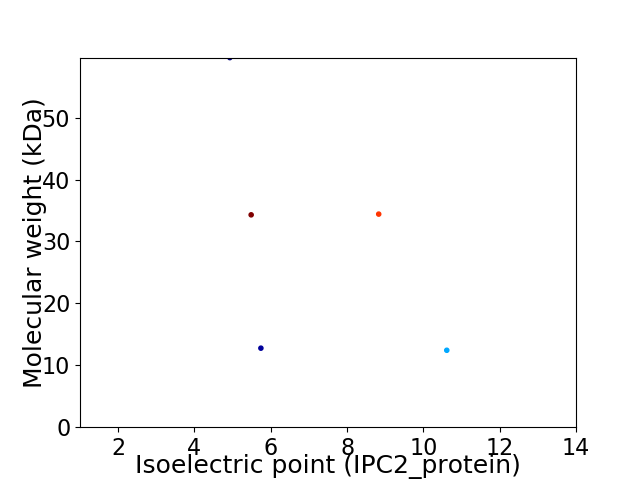
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
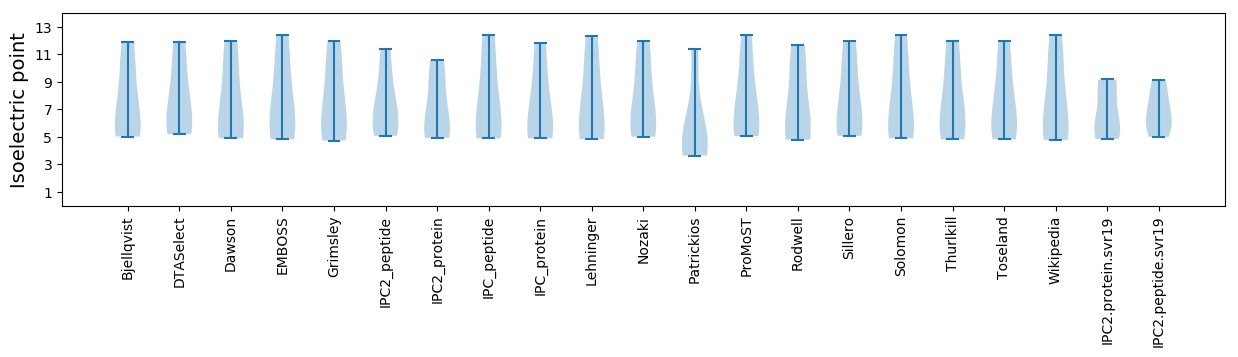
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UMH8|A0A0G2UMH8_9VIRU DNA pilot protein VP2 OS=Microviridae Fen7895_21 OX=1655660 PE=4 SV=1
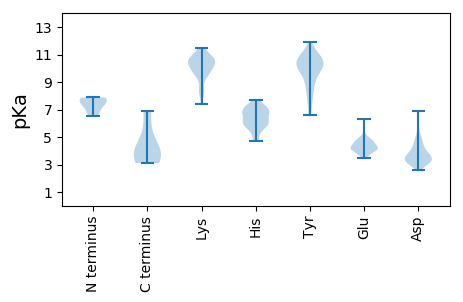
MM1 pKa = 7.19SQRR4 pKa = 11.84ILDD7 pKa = 3.99TGNKK11 pKa = 9.63ALVEE15 pKa = 4.1RR16 pKa = 11.84PVVNVPSNHH25 pKa = 5.92FNLSHH30 pKa = 7.11DD31 pKa = 4.03VKK33 pKa = 10.68LSCNIGQLIPCCTMEE48 pKa = 4.72MLPGDD53 pKa = 3.55RR54 pKa = 11.84VNINTEE60 pKa = 3.67ALVRR64 pKa = 11.84FQPMIAPIMHH74 pKa = 7.04RR75 pKa = 11.84LSIRR79 pKa = 11.84SEE81 pKa = 3.68WFFVPNRR88 pKa = 11.84LLWPKK93 pKa = 9.68WEE95 pKa = 4.82DD96 pKa = 3.99FISPPIEE103 pKa = 4.26GVEE106 pKa = 4.17PPAVPLIAEE115 pKa = 4.58TDD117 pKa = 3.21WSVTPNSLGDD127 pKa = 3.87YY128 pKa = 10.51LGLPLGLFDD137 pKa = 3.61EE138 pKa = 4.83TQPGVSALPFSAYY151 pKa = 8.98QRR153 pKa = 11.84IFYY156 pKa = 10.31DD157 pKa = 3.57WYY159 pKa = 10.35RR160 pKa = 11.84DD161 pKa = 3.12EE162 pKa = 6.33DD163 pKa = 4.69FYY165 pKa = 11.23PLQWQYY171 pKa = 11.91AGQLVDD177 pKa = 5.37GNNGDD182 pKa = 4.21NLDD185 pKa = 4.0EE186 pKa = 5.61LDD188 pKa = 4.35VIRR191 pKa = 11.84IRR193 pKa = 11.84NNDD196 pKa = 2.92RR197 pKa = 11.84DD198 pKa = 4.19YY199 pKa = 10.75FTSAKK204 pKa = 9.58PWAQKK209 pKa = 10.55GVVVTIPLLNTTLTLDD225 pKa = 3.31NAGYY229 pKa = 10.8AFSKK233 pKa = 10.82SDD235 pKa = 3.09GSAPNVGGAKK245 pKa = 9.91FDD247 pKa = 3.54GTGFQLEE254 pKa = 4.81DD255 pKa = 3.41VNGDD259 pKa = 3.65QLFLDD264 pKa = 4.0SPYY267 pKa = 11.0AITPEE272 pKa = 4.38DD273 pKa = 3.29ATAAAGTIEE282 pKa = 4.18QLRR285 pKa = 11.84QAVALQKK292 pKa = 10.62FLEE295 pKa = 4.53ADD297 pKa = 3.05ARR299 pKa = 11.84GGTRR303 pKa = 11.84YY304 pKa = 9.83VEE306 pKa = 4.79LLWQHH311 pKa = 5.77FEE313 pKa = 4.5EE314 pKa = 4.67YY315 pKa = 9.95MEE317 pKa = 5.13DD318 pKa = 3.86YY319 pKa = 10.49RR320 pKa = 11.84AQRR323 pKa = 11.84CEE325 pKa = 4.0YY326 pKa = 10.03IGNTVQPITISEE338 pKa = 4.27VLNTTGTDD346 pKa = 3.31AAPQGTMSGHH356 pKa = 6.69GISVARR362 pKa = 11.84HH363 pKa = 5.51ADD365 pKa = 3.0GLYY368 pKa = 10.03YY369 pKa = 9.99HH370 pKa = 7.12AKK372 pKa = 8.55EE373 pKa = 4.82HH374 pKa = 6.1GFLMCIMSVIPVSGYY389 pKa = 8.4YY390 pKa = 9.81QGVPKK395 pKa = 9.85MWSRR399 pKa = 11.84YY400 pKa = 9.38DD401 pKa = 3.5RR402 pKa = 11.84LDD404 pKa = 3.54YY405 pKa = 10.86AWPEE409 pKa = 3.87FAHH412 pKa = 6.93LGEE415 pKa = 3.93QAILNKK421 pKa = 9.77EE422 pKa = 4.0VYY424 pKa = 10.3YY425 pKa = 10.84DD426 pKa = 3.64VASTDD431 pKa = 3.4PVTGNEE437 pKa = 4.44GVWGYY442 pKa = 8.82VPRR445 pKa = 11.84YY446 pKa = 8.54SEE448 pKa = 4.69YY449 pKa = 10.49RR450 pKa = 11.84IQQNRR455 pKa = 11.84VSGDD459 pKa = 2.92FRR461 pKa = 11.84TSLDD465 pKa = 3.3YY466 pKa = 10.5WHH468 pKa = 7.71LARR471 pKa = 11.84KK472 pKa = 8.16FASLPPLAKK481 pKa = 10.19EE482 pKa = 4.35FLQISQGADD491 pKa = 2.9MDD493 pKa = 4.97RR494 pKa = 11.84IFAVIDD500 pKa = 3.32PTVQHH505 pKa = 6.45VLAHH509 pKa = 6.97WFHH512 pKa = 5.8QLKK515 pKa = 10.16FMRR518 pKa = 11.84KK519 pKa = 8.34LPKK522 pKa = 10.29LVVPSII528 pKa = 3.89
MM1 pKa = 7.19SQRR4 pKa = 11.84ILDD7 pKa = 3.99TGNKK11 pKa = 9.63ALVEE15 pKa = 4.1RR16 pKa = 11.84PVVNVPSNHH25 pKa = 5.92FNLSHH30 pKa = 7.11DD31 pKa = 4.03VKK33 pKa = 10.68LSCNIGQLIPCCTMEE48 pKa = 4.72MLPGDD53 pKa = 3.55RR54 pKa = 11.84VNINTEE60 pKa = 3.67ALVRR64 pKa = 11.84FQPMIAPIMHH74 pKa = 7.04RR75 pKa = 11.84LSIRR79 pKa = 11.84SEE81 pKa = 3.68WFFVPNRR88 pKa = 11.84LLWPKK93 pKa = 9.68WEE95 pKa = 4.82DD96 pKa = 3.99FISPPIEE103 pKa = 4.26GVEE106 pKa = 4.17PPAVPLIAEE115 pKa = 4.58TDD117 pKa = 3.21WSVTPNSLGDD127 pKa = 3.87YY128 pKa = 10.51LGLPLGLFDD137 pKa = 3.61EE138 pKa = 4.83TQPGVSALPFSAYY151 pKa = 8.98QRR153 pKa = 11.84IFYY156 pKa = 10.31DD157 pKa = 3.57WYY159 pKa = 10.35RR160 pKa = 11.84DD161 pKa = 3.12EE162 pKa = 6.33DD163 pKa = 4.69FYY165 pKa = 11.23PLQWQYY171 pKa = 11.91AGQLVDD177 pKa = 5.37GNNGDD182 pKa = 4.21NLDD185 pKa = 4.0EE186 pKa = 5.61LDD188 pKa = 4.35VIRR191 pKa = 11.84IRR193 pKa = 11.84NNDD196 pKa = 2.92RR197 pKa = 11.84DD198 pKa = 4.19YY199 pKa = 10.75FTSAKK204 pKa = 9.58PWAQKK209 pKa = 10.55GVVVTIPLLNTTLTLDD225 pKa = 3.31NAGYY229 pKa = 10.8AFSKK233 pKa = 10.82SDD235 pKa = 3.09GSAPNVGGAKK245 pKa = 9.91FDD247 pKa = 3.54GTGFQLEE254 pKa = 4.81DD255 pKa = 3.41VNGDD259 pKa = 3.65QLFLDD264 pKa = 4.0SPYY267 pKa = 11.0AITPEE272 pKa = 4.38DD273 pKa = 3.29ATAAAGTIEE282 pKa = 4.18QLRR285 pKa = 11.84QAVALQKK292 pKa = 10.62FLEE295 pKa = 4.53ADD297 pKa = 3.05ARR299 pKa = 11.84GGTRR303 pKa = 11.84YY304 pKa = 9.83VEE306 pKa = 4.79LLWQHH311 pKa = 5.77FEE313 pKa = 4.5EE314 pKa = 4.67YY315 pKa = 9.95MEE317 pKa = 5.13DD318 pKa = 3.86YY319 pKa = 10.49RR320 pKa = 11.84AQRR323 pKa = 11.84CEE325 pKa = 4.0YY326 pKa = 10.03IGNTVQPITISEE338 pKa = 4.27VLNTTGTDD346 pKa = 3.31AAPQGTMSGHH356 pKa = 6.69GISVARR362 pKa = 11.84HH363 pKa = 5.51ADD365 pKa = 3.0GLYY368 pKa = 10.03YY369 pKa = 9.99HH370 pKa = 7.12AKK372 pKa = 8.55EE373 pKa = 4.82HH374 pKa = 6.1GFLMCIMSVIPVSGYY389 pKa = 8.4YY390 pKa = 9.81QGVPKK395 pKa = 9.85MWSRR399 pKa = 11.84YY400 pKa = 9.38DD401 pKa = 3.5RR402 pKa = 11.84LDD404 pKa = 3.54YY405 pKa = 10.86AWPEE409 pKa = 3.87FAHH412 pKa = 6.93LGEE415 pKa = 3.93QAILNKK421 pKa = 9.77EE422 pKa = 4.0VYY424 pKa = 10.3YY425 pKa = 10.84DD426 pKa = 3.64VASTDD431 pKa = 3.4PVTGNEE437 pKa = 4.44GVWGYY442 pKa = 8.82VPRR445 pKa = 11.84YY446 pKa = 8.54SEE448 pKa = 4.69YY449 pKa = 10.49RR450 pKa = 11.84IQQNRR455 pKa = 11.84VSGDD459 pKa = 2.92FRR461 pKa = 11.84TSLDD465 pKa = 3.3YY466 pKa = 10.5WHH468 pKa = 7.71LARR471 pKa = 11.84KK472 pKa = 8.16FASLPPLAKK481 pKa = 10.19EE482 pKa = 4.35FLQISQGADD491 pKa = 2.9MDD493 pKa = 4.97RR494 pKa = 11.84IFAVIDD500 pKa = 3.32PTVQHH505 pKa = 6.45VLAHH509 pKa = 6.97WFHH512 pKa = 5.8QLKK515 pKa = 10.16FMRR518 pKa = 11.84KK519 pKa = 8.34LPKK522 pKa = 10.29LVVPSII528 pKa = 3.89
Molecular weight: 59.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UK59|A0A0G2UK59_9VIRU Major capsid protein VP1 OS=Microviridae Fen7895_21 OX=1655660 PE=3 SV=1
MM1 pKa = 7.73IMLNLFRR8 pKa = 11.84ISLIGSLLLYY18 pKa = 9.65LIRR21 pKa = 11.84YY22 pKa = 7.84FPFGRR27 pKa = 11.84LLIVIRR33 pKa = 11.84VGRR36 pKa = 11.84LCLLLVRR43 pKa = 11.84LTLAICLSIPLISIMLISSISLLLLVLLVRR73 pKa = 11.84ALNLLMMSLRR83 pKa = 11.84YY84 pKa = 9.65VKK86 pKa = 10.28QLLLSIRR93 pKa = 11.84LKK95 pKa = 10.89RR96 pKa = 11.84PLTVLSLKK104 pKa = 10.43KK105 pKa = 10.31RR106 pKa = 11.84LLL108 pKa = 3.79
MM1 pKa = 7.73IMLNLFRR8 pKa = 11.84ISLIGSLLLYY18 pKa = 9.65LIRR21 pKa = 11.84YY22 pKa = 7.84FPFGRR27 pKa = 11.84LLIVIRR33 pKa = 11.84VGRR36 pKa = 11.84LCLLLVRR43 pKa = 11.84LTLAICLSIPLISIMLISSISLLLLVLLVRR73 pKa = 11.84ALNLLMMSLRR83 pKa = 11.84YY84 pKa = 9.65VKK86 pKa = 10.28QLLLSIRR93 pKa = 11.84LKK95 pKa = 10.89RR96 pKa = 11.84PLTVLSLKK104 pKa = 10.43KK105 pKa = 10.31RR106 pKa = 11.84LLL108 pKa = 3.79
Molecular weight: 12.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1372 |
108 |
528 |
274.4 |
30.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.184 ± 1.948 | 1.02 ± 0.369 |
5.977 ± 0.905 | 3.644 ± 0.827 |
3.863 ± 0.826 | 5.394 ± 0.787 |
2.332 ± 0.604 | 5.685 ± 0.76 |
4.009 ± 0.459 | 10.204 ± 2.715 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.915 ± 0.629 | 6.122 ± 1.344 |
5.539 ± 0.714 | 5.102 ± 0.89 |
5.685 ± 1.201 | 5.758 ± 0.467 |
5.758 ± 1.105 | 6.778 ± 0.493 |
1.312 ± 0.544 | 3.717 ± 0.94 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
